| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:34:35 UTC |
|---|
| Update Date | 2016-11-09 01:22:30 UTC |
|---|
| Accession Number | CHEM041575 |
|---|
| Identification |
|---|
| Common Name | Caffeic acid 3-O-glucuronide |
|---|
| Class | Small Molecule |
|---|
| Description | Caffeic acid 3-O-glucuronide is a polyphenol metabolite detected in biological fluids (PMID: 20428313). |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
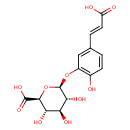
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Caffeate 3-O-glucuronide | Generator | | (2S,3S,4S,5R,6S)-6-{5-[(1E)-2-carboxyeth-1-en-1-yl]-2-hydroxyphenoxy}-3,4,5-trihydroxyoxane-2-carboxylate | HMDB |
|
|---|
| Chemical Formula | C15H16O10 |
|---|
| Average Molecular Mass | 356.282 g/mol |
|---|
| Monoisotopic Mass | 356.074 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2S,3S,4S,5R,6S)-6-{5-[(1E)-2-carboxyeth-1-en-1-yl]-2-hydroxyphenoxy}-3,4,5-trihydroxyoxane-2-carboxylic acid |
|---|
| Traditional Name | (2S,3S,4S,5R,6S)-6-{5-[(1E)-2-carboxyeth-1-en-1-yl]-2-hydroxyphenoxy}-3,4,5-trihydroxyoxane-2-carboxylic acid |
|---|
| SMILES | O[C@H]1[C@H](OC2=C(O)C=CC(\C=C\C(O)=O)=C2)O[C@@H]([C@@H](O)[C@@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C15H16O10/c16-7-3-1-6(2-4-9(17)18)5-8(7)24-15-12(21)10(19)11(20)13(25-15)14(22)23/h1-5,10-13,15-16,19-21H,(H,17,18)(H,22,23)/b4-2+/t10-,11-,12+,13-,15+/m0/s1 |
|---|
| InChI Key | BSOMSDFTZKNUHY-ZYZFHZPYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Phenolic glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenolic glycoside
- 1-o-glucuronide
- O-glucuronide
- Glucuronic acid or derivatives
- Cinnamic acid
- Cinnamic acid or derivatives
- Coumaric acid or derivatives
- Hexose monosaccharide
- Hydroxycinnamic acid
- Hydroxycinnamic acid or derivatives
- O-glycosyl compound
- Phenol ether
- Styrene
- Phenoxy compound
- Phenol
- 1-hydroxy-2-unsubstituted benzenoid
- Beta-hydroxy acid
- Pyran
- Benzenoid
- Oxane
- Monosaccharide
- Hydroxy acid
- Monocyclic benzene moiety
- Dicarboxylic acid or derivatives
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Carboxylic acid derivative
- Carboxylic acid
- Acetal
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052r-9354000000-a1df090612e7ece45584 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00c0-2601149000-7399b5b3aa0cfc21a13a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q0-0819000000-7f76e3b3d77753808a27 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-0901000000-dffbcafd6d039934c09a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0089-1900000000-63b5f2d4b2246aecbe46 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0bvi-1619000000-d29bc89d06ae25debde3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-1912000000-e4798d2f7dba480720a9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-2900000000-060c19e93edb734a81dd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dr-0906000000-3d12ab9c402a93454ebd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dr-0911000000-90e3e1c0bb702fd7ece6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kb-0900000000-f379172c6d69fd66e436 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0819000000-479ec1337f224b453ad5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-3921000000-93e51acc03012c33dec0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1910000000-c6e8fb2523ce090502db | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041705 |
|---|
| FooDB ID | FDB029870 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 30777604 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 25171992 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D, Scalbert A: Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford). 2010;2010:bap024. doi: 10.1093/database/bap024. Epub 2010 Jan 8. |
|
|---|