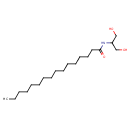
Palmitoyl serinol (CHEM041410)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-05-27 01:28:21 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2016-11-09 01:22:28 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | CHEM041410 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Palmitoyl serinol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 2-Palmitoyl glycerol (2-PG) has been isolated along with the potent endocannabinoid 2-arachidonoyl glycerol (2-AG) from various tissues.1 Although 2-PG displays no intrinsic agonist activity on CB1 or CB2 receptors, it does potentiate the ability of 2-AG to inhibit adenylyl cyclase. 2-PG also potentiates the analgesic, hypokinetic, and anxiolytic effects of 2-AG in mice. This "entourage" effect has been attributed to the ability of compounds such as 2-PG to inhibit reuptake and/or compete with the active endocannabinoids for access to inactivating enzymes such as FAAH and monoglyceride lipase.2,3 Palmitoyl serinol is a stable analog of 2-PG bearing an amide linkage in place of the labile glyceryl ester. This has the potential to enhance its "entourage" activities as a result of a prolonged in vivo half-life. Palmitoyl serinol is also an analog of C-16 ceramide. Incubation of neuroblastoma cells with palmitoyl serinol causes apoptosis with an IC50 of approximately 80 µM. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Contaminant Sources |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Contaminant Type | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C19H39NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 329.518 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 329.293 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 126127-31-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | N-(1,3-dihydroxypropan-2-yl)hexadecanamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | N-(1,3-dihydroxypropan-2-yl)hexadecanamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CCCCCCCCCCCCCCCC(=O)NC(CO)CO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C19H39NO3/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-19(23)20-18(16-21)17-22/h18,21-22H,2-17H2,1H3,(H,20,23) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | MZUNFYMZKTWADX-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as n-acyl amines. N-acyl amines are compounds containing a fatty acid moiety linked to an amine group through an ester linkage. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty amides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | N-acyl amines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0013654 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB029624 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 8038003 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 9862307 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kegg Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| YMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ECMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||