| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:19:06 UTC |
|---|
| Update Date | 2016-11-09 01:22:26 UTC |
|---|
| Accession Number | CHEM041267 |
|---|
| Identification |
|---|
| Common Name | PGP(16:0/16:0) |
|---|
| Class | Small Molecule |
|---|
| Description | A 3-(3-sn-phosphatidyl)-sn-glycerol 1-phosphate in which both phosphatidyl acyl groups are specified as hexadecanoyl (palmitoyl). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
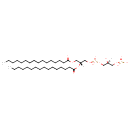
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Dihexadecanoylphosphatidylglycerol phosphate | ChEBI | | 1,2-Dipalmitoyl-sn-glycero-3-phospho-(1'-sn-glycerol-3'-phosphate) | ChEBI | | 1,2-Dipalmitoylphosphatidylglycerol phosphate | ChEBI | | Phosphatidylglycerol phosphate 16:0/16:0 | ChEBI | | 1,2-Dihexadecanoylphosphatidylglycerol phosphoric acid | Generator | | 1,2-Dipalmitoyl-sn-glycero-3-phospho-(1'-sn-glycerol-3'-phosphoric acid) | Generator | | 1,2-Dipalmitoylphosphatidylglycerol phosphoric acid | Generator | | Phosphatidylglycerol phosphoric acid 16:0/16:0 | Generator | | 1,2-Dipalmitoyl-rac-glycero-3-phospho-(1'-sn-glycerol-3'-phosphate) | HMDB | | 3-sn-Phosphatidyl-1'-sn-glycerol 3'-phosphoric acid | HMDB | | PGP(32:0) | HMDB | | 1,2-Dihexadecanoyl-rac-glycero-3-phospho-(1'-sn-glycerol-3'-phosphate) | HMDB | | PGP(16:0/16:0) | Lipid Annotator |
|
|---|
| Chemical Formula | C38H76O13P2 |
|---|
| Average Molecular Mass | 802.961 g/mol |
|---|
| Monoisotopic Mass | 802.476 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | [(2S)-3-({[(2R)-2,3-bis(hexadecanoyloxy)propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphonic acid |
|---|
| Traditional Name | (2S)-3-{[(2R)-2,3-bis(hexadecanoyloxy)propoxy(hydroxy)phosphoryl]oxy}-2-hydroxypropoxyphosphonic acid |
|---|
| SMILES | [H][C@](O)(COP(O)(O)=O)COP(O)(=O)OC[C@@]([H])(COC(=O)CCCCCCCCCCCCCCC)OC(=O)CCCCCCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C38H76O13P2/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-37(40)47-33-36(34-50-53(45,46)49-32-35(39)31-48-52(42,43)44)51-38(41)30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h35-36,39H,3-34H2,1-2H3,(H,45,46)(H2,42,43,44)/t35-,36+/m0/s1 |
|---|
| InChI Key | ONJBJMDJKLHMEK-MPQUPPDSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylglycerophosphates. These are glycerophosphoglycerophosphates in which two fatty acids are bonded to the 1-glycerol moiety through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoglycerophosphates |
|---|
| Direct Parent | Phosphatidylglycerophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diacylglycerophosphoglycerophosphate
- Sn-glycerol-3-phosphate
- Fatty acid ester
- Monoalkyl phosphate
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Secondary alcohol
- Carboxylic acid ester
- Carboxylic acid derivative
- Organooxygen compound
- Organic oxide
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0k9j-0480191630-8f5da3dc73589ad27ea3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052s-1592041300-161f80080f7f88795617 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4s-4963241100-06672c7b8e06515f0d96 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0pds-3090031020-4b194687790d63ffb8ef | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9050000000-a1bf46fc4f76cc8acee4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9010000000-168aacc7c520fbf633d2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0010000090-5619e2fc6f74a1450058 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-3091041050-9cf6e68eaceac0f75069 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-5093082100-737807923db4f959eef0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1210060590-6c187fc092c84c325aa1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0pb9-3700196400-ef4a3a2a41f9662f5a04 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udj-4395600000-e7daddc00075edaa1f13 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0013472 |
|---|
| FooDB ID | FDB029472 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 136773 |
|---|
| PubChem Compound ID | 49859598 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | YMDB14372 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|