| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:17:42 UTC |
|---|
| Update Date | 2016-11-09 01:22:26 UTC |
|---|
| Accession Number | CHEM041248 |
|---|
| Identification |
|---|
| Common Name | PC(o-22:1(13Z)/22:2(13Z,16Z)) |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
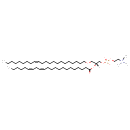
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Erucyl-2-docosadienoyl-sn-glycero-3-phosphocholine | HMDB | | Gpcho(22:1/22:2) | HMDB | | Gpcho(22:1n9/22:2n6) | HMDB | | Gpcho(22:1W9/22:2W6) | HMDB | | Gpcho(44:3) | HMDB | | Lecithin | HMDB | | PC Ae C44:3 | HMDB | | PC(22:1/22:2) | HMDB | | PC(22:1n9/22:2n6) | HMDB | | PC(22:1W9/22:2W6) | HMDB | | PC(44:3) | HMDB | | PC(O-44:3) | HMDB | | Phosphatidylcholine(22:1/22:2) | HMDB | | Phosphatidylcholine(22:1n9/22:2n6) | HMDB | | Phosphatidylcholine(22:1W9/22:2W6) | HMDB | | Phosphatidylcholine(44:3) | HMDB | | 1-(13Z-Docosenyl)-2-(13Z,16Z-docosadienoyl)-sn-glycero-3-phosphocholine | HMDB | | PC(o-22:1(13Z)/22:2(13Z,16Z)) | Lipid Annotator |
|
|---|
| Chemical Formula | C52H100NO7P |
|---|
| Average Molecular Mass | 882.327 g/mol |
|---|
| Monoisotopic Mass | 881.724 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2-{[(2R)-3-[(13Z)-docos-13-en-1-yloxy]-2-[(13Z,16Z)-docosa-13,16-dienoyloxy]propyl phosphonato]oxy}ethyl)trimethylazanium |
|---|
| Traditional Name | (2-{[(2R)-3-[(13Z)-docos-13-en-1-yloxy]-2-[(13Z,16Z)-docosa-13,16-dienoyloxy]propyl phosphonato]oxy}ethyl)trimethylazanium |
|---|
| SMILES | CCCCCCCC\C=C/CCCCCCCCCCCCOC[C@]([H])(COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CCCCCCCCCCC\C=C/C\C=C/CCCCC |
|---|
| InChI Identifier | InChI=1S/C52H100NO7P/c1-6-8-10-12-14-16-18-20-22-24-26-28-30-32-34-36-38-40-42-44-47-57-49-51(50-59-61(55,56)58-48-46-53(3,4)5)60-52(54)45-43-41-39-37-35-33-31-29-27-25-23-21-19-17-15-13-11-9-7-2/h15,17,20-23,51H,6-14,16,18-19,24-50H2,1-5H3/b17-15-,22-20-,23-21-/t51-/m1/s1 |
|---|
| InChI Key | VCODWVLGDPOACA-DRGSQYEJSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-alkyl,2-acylglycero-3-phosphocholines. These are glycerophosphocholines that carry exactly one acyl chain attached to the glycerol moiety through an ester linkage at the O2-position, and one alkyl chain attached through an ether linkage at the O1-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | 1-alkyl,2-acylglycero-3-phosphocholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-alkyl,2-acylglycero-3-phosphocholine
- Phosphocholine
- Fatty acid ester
- Dialkyl phosphate
- Glycerol ether
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Fatty acyl
- Alkyl phosphate
- Tetraalkylammonium salt
- Quaternary ammonium salt
- Carboxylic acid ester
- Carboxylic acid derivative
- Dialkyl ether
- Ether
- Monocarboxylic acid or derivatives
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Amine
- Organic nitrogen compound
- Organooxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organic salt
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-9105031230-8ac1e5f24a5c527f32e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0cei-5309021210-42e5c8d6c14d4675f85e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05nr-9018003110-46c70a6ed4470cd911fb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001r-0007000290-3f3a5f0be83f7ebcdf88 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bi-2019200630-ae7ecbfd4f323ccdc68a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05p9-4019200000-aa4951f16661fed11de0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000090-96e9e570336d9ee74713 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0000000090-96e9e570336d9ee74713 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-1900061070-5d86dc6afcf6786a42aa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000000009-ca3f7b3580cfc8035037 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0000000009-ca3f7b3580cfc8035037 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4r-0007120191-89d350907d4d2fe5cbf7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000000090-bd09935a9e14a7e78d8f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00lr-0005001090-79db4730e166c2c0d5bd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-002r-4009400000-55c8e25f625487b4c186 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0013452 |
|---|
| FooDB ID | FDB029452 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 89997 |
|---|
| PubChem Compound ID | 53481759 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|