| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:14:49 UTC |
|---|
| Update Date | 2016-11-09 01:22:24 UTC |
|---|
| Accession Number | CHEM041123 |
|---|
| Identification |
|---|
| Common Name | 18-Hydroxy-11-dehydrotetrahydrocorticosterone |
|---|
| Class | Small Molecule |
|---|
| Description | A human metabolite taken as a putative food compound of mammalian origin |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
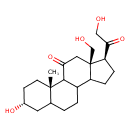
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 18-Hydroxy-11-dehydrotetrahydrocorticosterone | MeSH | | 18-Hydroxytetrahydro-11-dehydrocorticosterone | MeSH | | HTDHC | MeSH |
|
|---|
| Chemical Formula | C21H32O5 |
|---|
| Average Molecular Mass | 364.476 g/mol |
|---|
| Monoisotopic Mass | 364.225 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2S,5R,14S,15R)-5-hydroxy-14-(2-hydroxyacetyl)-15-(hydroxymethyl)-2-methyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-17-one |
|---|
| Traditional Name | (2S,5R,14S,15R)-5-hydroxy-14-(2-hydroxyacetyl)-15-(hydroxymethyl)-2-methyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-17-one |
|---|
| SMILES | C[C@]12CC[C@@H](O)CC1CCC1C3CC[C@H](C(=O)CO)[C@@]3(CO)CC(=O)C21 |
|---|
| InChI Identifier | InChI=1S/C21H32O5/c1-20-7-6-13(24)8-12(20)2-3-14-15-4-5-16(18(26)10-22)21(15,11-23)9-17(25)19(14)20/h12-16,19,22-24H,2-11H2,1H3/t12?,13-,14?,15?,16-,19?,20+,21-/m1/s1 |
|---|
| InChI Key | UQRPWWKJWZFNAE-HATFOLNYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 21-hydroxysteroids. These are steroids carrying a hydroxyl group at the 21-position of the steroid backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Hydroxysteroids |
|---|
| Direct Parent | 21-hydroxysteroids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Progestogin-skeleton
- 21-hydroxysteroid
- 20-oxosteroid
- Pregnane-skeleton
- 18-hydroxysteroid
- 3-hydroxysteroid
- 11-oxosteroid
- Oxosteroid
- 3-alpha-hydroxysteroid
- Cyclic alcohol
- Alpha-hydroxy ketone
- Ketone
- Secondary alcohol
- Alcohol
- Hydrocarbon derivative
- Organooxygen compound
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Primary alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05v0-0309000000-e5a3a529d17a1914a71d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-014i-1100390000-44bed125b167e579eb3e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00mk-0009000000-074b3045e311f290f057 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00os-0019000000-7d5473b172f18a3846e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kr-3298000000-1fff7b1e86db6bfc8bec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-75719bdc77ba082a441c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-07wa-1009000000-37c9b0c0f503fed120d7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-4096000000-05e7ac1fcc9232d81363 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-2d1f7ed49e286f1be540 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9028000000-4c21c514e3ea3fa3b9f7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01wf-3059000000-576b1375b2407cb4a579 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0019000000-3e5f07ce2d4cbfb24861 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00mk-0129000000-9a3fb1d7ed97210fbd1e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-9742000000-b862d3ba40afe083bcdf | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0013157 |
|---|
| FooDB ID | FDB029313 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53481635 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|