| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:13:39 UTC |
|---|
| Update Date | 2016-11-09 01:22:24 UTC |
|---|
| Accession Number | CHEM041105 |
|---|
| Identification |
|---|
| Common Name | Xanthurenate-8-O-beta-D-glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
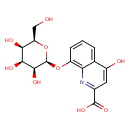
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Xanthurenate-8-O-b-D-glucoside | Generator | | Xanthurenate-8-O-β-D-glucoside | Generator | | Xanthurenic acid-8-O-b-D-glucoside | Generator | | Xanthurenic acid-8-O-beta-D-glucoside | Generator | | Xanthurenic acid-8-O-β-D-glucoside | Generator | | 4,8-Dihydroxy-8-O-beta-D-glucoside-quinaldic acid anion | HMDB | | 4,8-Dihydroxy-8-O-beta-D-glucoside-quinoline-2-carboxylic acid anion | HMDB | | 4,8-Dihydroxy-8-O-beta-delta-glucoside-quinaldic acid anion | HMDB | | 4,8-Dihydroxy-8-O-beta-delta-glucoside-quinoline-2-carboxylic acid anion | HMDB | | 4,8-Dihydroxyquinaldic acid 8-O-beta-D-glucoside | HMDB | | 4,8-Dihydroxyquinaldic acid 8-O-beta-delta-glucoside | HMDB | | 4,8-Dihydroxyquinoline-2-carboxylic acid 8-O-beta-D-glucoside | HMDB | | 4,8-Dihydroxyquinoline-2-carboxylic acid 8-O-beta-delta-glucoside | HMDB | | Xan8GLC | HMDB | | Xanthurenic acid 8-O-beta-D-glucoside | HMDB | | Xanthurenic acid 8-O-beta-delta-glucoside | HMDB | | Xanthurenic acid 8-O-glucoside | MeSH, HMDB | | Cardinalic acid | MeSH, HMDB | | 4-Hydroxy-8-{[(2S,3S,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}quinoline-2-carboxylate | Generator |
|
|---|
| Chemical Formula | C16H17NO9 |
|---|
| Average Molecular Mass | 367.308 g/mol |
|---|
| Monoisotopic Mass | 367.090 g/mol |
|---|
| CAS Registry Number | 97451-32-6 |
|---|
| IUPAC Name | 4-hydroxy-8-{[(2S,3S,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}quinoline-2-carboxylic acid |
|---|
| Traditional Name | 4-hydroxy-8-{[(2S,3S,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}quinoline-2-carboxylic acid |
|---|
| SMILES | OC[C@H]1O[C@@H](OC2=C3N=C(C=C(O)C3=CC=C2)C(O)=O)[C@@H](O)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C16H17NO9/c18-5-10-12(20)13(21)14(22)16(26-10)25-9-3-1-2-6-8(19)4-7(15(23)24)17-11(6)9/h1-4,10,12-14,16,18,20-22H,5H2,(H,17,19)(H,23,24)/t10-,12+,13+,14+,16-/m1/s1 |
|---|
| InChI Key | MYFHOUJDPFBJLH-XGJKELJWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Phenolic glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenolic glycoside
- Quinoline-2-carboxylic acid
- Hexose monosaccharide
- Dihydroquinolone
- O-glycosyl compound
- Dihydroquinoline
- Quinoline
- Pyridine carboxylic acid
- Pyridine carboxylic acid or derivatives
- Monosaccharide
- Oxane
- Benzenoid
- Pyridine
- Heteroaromatic compound
- Vinylogous amide
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Oxacycle
- Azacycle
- Polyol
- Organoheterocyclic compound
- Alcohol
- Hydrocarbon derivative
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Primary alcohol
- Organic oxide
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uds-5926000000-404b2e8d5851ea309efd | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0006-7332039000-350d78b17196c6d21ed5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gb9-0249000000-3063a43361b169d1e41a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1896000000-d0d6f5f037cf706f3de1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-114l-3940000000-01e636ec4d1c9981a108 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0pvi-0259000000-8da58724f8100d50efd5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bti-0971000000-bd91a20ae5011a44f09b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06tr-2930000000-d907e683d10b2b2f124f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0219000000-18106fd97aab020dc8c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kmi-1339000000-5d4431e369fbcf176722 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06r2-8950000000-8dbbaa9cf45b63a28a26 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0v4i-0109000000-919c5c092ea5bb4ed547 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0r00-2945000000-04f0431dc9dbf1f6e460 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0btc-3920000000-43ce5a860e4fac9ebe25 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0013118 |
|---|
| FooDB ID | FDB029293 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 30776690 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53481609 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|