| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:11:16 UTC |
|---|
| Update Date | 2016-11-09 01:22:23 UTC |
|---|
| Accession Number | CHEM041078 |
|---|
| Identification |
|---|
| Common Name | Pentanoyl-CoA |
|---|
| Class | Small Molecule |
|---|
| Description | Pentanoyl coA is a acyl-CoA with the C-5 Acyl chain.
Acyl-CoA (or formyl-CoA) is a coenzyme involved in the metabolism of fatty acids. It is a temporary compound formed when coenzyme A (CoA) attaches to the end of a long-chain fatty acid, inside living cells. The CoA is then removed from the chain, carrying two carbons from the chain with it, forming acetyl-CoA. This is then used in the citric acid cycle to start a chain of reactions, eventually forming many adenosine triphosphates.
To be oxidatively degraded, a fatty acid must first be activated in a two-step reaction catalyzed by acyl-CoA synthetase. First, the fatty acid displaces the diphosphate group of ATP, then coenzyme A (HSCoA) displaces the AMP group to form an Acyl-CoA. The acyladenylate product of the first step has a large free energy of hydrolysis and conserves the free energy of the cleaved phosphoanhydride bond in ATP. The second step, transfer of the acyl group to CoA (the same molecule that carries acetyl groups as acetyl-CoA), conserves free energy in the formation of a thioester bond. Consequently, the overall reaction
Fatty acid + CoA + ATP <=> Acyl-CoA + AMP + PPi
has a free energy change near zero. Subsequent hydrolysis of the product PPi (by the enzyme inorganic pyrophosphatase) is highly exergonic, and this reaction makes the formation of acyl-CoA spontaneous and irreversible.
Fatty acids are activated in the cytosol, but oxidation occurs in the mitochondria. Because there is no transport protein for CoA adducts, acyl groups must enter the mitochondria via a shuttle system involving the small molecule carnitine. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
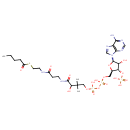
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Pentanoyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C26H44N7O17P3S |
|---|
| Average Molecular Mass | 851.651 g/mol |
|---|
| Monoisotopic Mass | 851.173 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy({3-hydroxy-2,2-dimethyl-3-[(2-{[2-(pentanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy})phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-({[hydroxy([hydroxy(3-hydroxy-2,2-dimethyl-3-[(2-{[2-(pentanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy)phosphoryl]oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxyphosphonic acid |
|---|
| SMILES | CCCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C26H44N7O17P3S/c1-4-5-6-17(35)54-10-9-28-16(34)7-8-29-24(38)21(37)26(2,3)12-47-53(44,45)50-52(42,43)46-11-15-20(49-51(39,40)41)19(36)25(48-15)33-14-32-18-22(27)30-13-31-23(18)33/h13-15,19-21,25,36-37H,4-12H2,1-3H3,(H,28,34)(H,29,38)(H,42,43)(H,44,45)(H2,27,30,31)(H2,39,40,41)/t15-,19-,20-,21?,25-/m1/s1 |
|---|
| InChI Key | RXUATCUKICAIOA-TVCSPYKZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of inorganic compounds known as homogeneous transition metal compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a transition metal atom. |
|---|
| Kingdom | Inorganic compounds |
|---|
| Super Class | Homogeneous metal compounds |
|---|
| Class | Homogeneous transition metal compounds |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Homogeneous transition metal compounds |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | - Homogeneous transition metal
|
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-2901000120-2a423c1bd81049be2f37 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-1913000000-a7882da7f4659358dd99 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1911000000-dd4d2426d295e6cea710 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-7930140450-c93bb9584e9108db8543 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-6910010000-390528f5d6ef7539cb68 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-5900100000-f9c4115d1b808c86f8f7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000090-3758b512fc2398854707 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900001560-656412ccb7e988988a2b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-0029000000-298400882ff5c02d176c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0000000090-c1d8a32a3fcbde2b3b71 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fas-9200202360-645cab450d162098ff13 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00p0-6204702920-60948d54a96619a917a8 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0013037 |
|---|
| FooDB ID | FDB029262 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | 5-HYDROXYPENTANOYL-COA |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 388465 |
|---|
| ChEBI ID | 15536 |
|---|
| PubChem Compound ID | 439337 |
|---|
| Kegg Compound ID | C00888 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|