| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:07:37 UTC |
|---|
| Update Date | 2016-11-09 01:22:22 UTC |
|---|
| Accession Number | CHEM040995 |
|---|
| Identification |
|---|
| Common Name | 4-Oxo-13-cis-retinoate |
|---|
| Class | Small Molecule |
|---|
| Description | 4-oxo-9-cis retinoic acid (4-oxo-9cRA) is identified as a major plasma metabolite of 9-cis retinoic acid. Plasma levels of 4-oxo-9-cRA were initially 71% of those of 9cRA, but in contrast to 9cRA, there was no decline in plasma levels.Despite a decline in plasma levels of 9cRA over time, levels of the 4-oxo metabolite tended to persist. While the 4-oxo metabolite is less potent than the parent compound. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
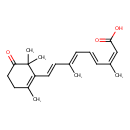
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-oxo-13-cis-Retinoic acid | Generator | | 3,7-Dimethyl-9-(5-oxo-2,6,6-trimethyl-1-cyclohexenyl)-(2E,4Z,6Z,8Z)-nonatetraen-1-Oate | HMDB | | 3,7-Dimethyl-9-(5-oxo-2,6,6-trimethyl-1-cyclohexenyl)-(2E,4Z,6Z,8Z)-nonatetraen-1-Oic acid | HMDB | | 4-keto-13-cis-Retinoate | HMDB | | 4-keto-13-cis-Retinoic acid | HMDB | | 4-oxo-13-cis-Retinoic acid anion | HMDB | | 4O13CVA | HMDB | | (2Z,4E,6E,8E)-3,7-Dimethyl-9-(2,6,6-trimethyl-5-oxocyclohex-1-en-1-yl)nona-2,4,6,8-tetraenoate | Generator |
|
|---|
| Chemical Formula | C20H26O3 |
|---|
| Average Molecular Mass | 314.419 g/mol |
|---|
| Monoisotopic Mass | 314.188 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2Z,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-5-oxocyclohex-1-en-1-yl)nona-2,4,6,8-tetraenoic acid |
|---|
| Traditional Name | (2Z,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-5-oxocyclohex-1-en-1-yl)nona-2,4,6,8-tetraenoic acid |
|---|
| SMILES | C\C(\C=C\C1=C(C)CCC(=O)C1(C)C)=C/C=C/C(/C)=C\C(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H26O3/c1-14(7-6-8-15(2)13-19(22)23)9-11-17-16(3)10-12-18(21)20(17,4)5/h6-9,11,13H,10,12H2,1-5H3,(H,22,23)/b8-6+,11-9+,14-7+,15-13- |
|---|
| InChI Key | YOFBBAJVETYSCH-IWRFDTMXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as retinoids. These are oxygenated derivatives of 3,7-dimethyl-1-(2,6,6-trimethylcyclohex-1-enyl)nona-1,3,5,7-tetraene and derivatives thereof. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Retinoids |
|---|
| Direct Parent | Retinoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Retinoic acid
- Diterpenoid
- Retinoid skeleton
- Medium-chain fatty acid
- Cyclohexenone
- Methyl-branched fatty acid
- Branched fatty acid
- Fatty acyl
- Fatty acid
- Unsaturated fatty acid
- Ketone
- Cyclic ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organooxygen compound
- Organic oxide
- Carbonyl group
- Hydrocarbon derivative
- Organic oxygen compound
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-2090000000-1c5451b849e980e311b0 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-3029000000-b04d7c59fc3ce4973812 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kb-0391000000-8caaf7c4c017bddfdaa6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-1980000000-af3a5fdeff98e8840ec4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kbk-4910000000-1506f3079c815eac1226 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03xr-0089000000-7665fb8471756952e98c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-02ta-0094000000-c917b32286fab5a78c61 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uxv-6290000000-55b32ed2f34d92f67cfa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0079000000-16a7e691d9676e7c84c5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0gb9-0290000000-83e5e9702daddae492f7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-016r-4910000000-e94b1001e037759106b0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0392000000-83a2fc1baaa988e42e55 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fbj-0490000000-53eac69477af49e3a40d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-4910000000-14c7b29334f7c8138747 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012789 |
|---|
| FooDB ID | FDB029175 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 8171361 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 9995780 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|