| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:05:51 UTC |
|---|
| Update Date | 2016-11-09 01:22:22 UTC |
|---|
| Accession Number | CHEM040963 |
|---|
| Identification |
|---|
| Common Name | 16E-18-Oxo-18-CoA-dinor-LTE4 |
|---|
| Class | Small Molecule |
|---|
| Description | 16E-18-oxo-18-CoA-dinor-LTE4 is a metabolite through lipid oxidation of Leukotriene E4 (LTE4).Leukotriene E4 (LTE4) is a cysteinyl leukotriene. Cysteinyl leukotrienes (CysLTs) are a family of potent inflammatory mediators that appear to contribute to the pathophysiologic features of allergic rhinitis. Nasal blockage induced by CysLTs is mainly due to dilatation of nasal blood vessels, which can be induced by the nitric oxide produced through CysLT1 receptor activation. LTE4, activate contractile and inflammatory processes via specific interaction with putative seven transmembrane-spanning receptors that couple to G proteins and subsequent intracellular signaling pathways. LTE4 is metabolized from leukotriene C4 in a reaction catalyzed by gamma-glutamyl transpeptidase and a particulate dipeptidase from kidney. (PMID: 12607939, 12432945, 6311078). Leukotrienes are eicosanoids. The eicosanoids consist of the prostaglandins (PGs), thromboxanes (TXs), leukotrienes (LTs), and lipoxins (LXs). The PGs and TXs are collectively identified as prostanoids. Prostaglandins were originally shown to be synthesized in the prostate gland, thromboxanes from platelets (thrombocytes), and leukotrienes from leukocytes, hence the derivation of their names. All mammalian cells except erythrocytes synthesize eicosanoids. These molecules are extremely potent, able to cause profound physiological effects at very dilute concentrations. All eicosanoids function locally at the site of synthesis, through receptor-mediated G-protein linked signalling pathways. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
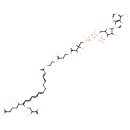
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| CoA-18-COOH-17E-Dinor-lte(,3) | HMDB | | coenzyme A-18-COOH-17E-Dinor-lte(,3) | HMDB | | 6-[(2-Amino-2-carboxyethyl)sulfanyl]-18-({2-[(3-{[4-({[({[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-1,2-dihydroxy-3,3-dimethylbutylidene]amino}-1-hydroxypropylidene)amino]ethyl}sulfanyl)-5-hydroxy-18-oxooctadeca-7,9,11,16-tetraenoate | HMDB | | 6-[(2-Amino-2-carboxyethyl)sulphanyl]-18-({2-[(3-{[4-({[({[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-1,2-dihydroxy-3,3-dimethylbutylidene]amino}-1-hydroxypropylidene)amino]ethyl}sulphanyl)-5-hydroxy-18-oxooctadeca-7,9,11,16-tetraenoate | HMDB | | 6-[(2-Amino-2-carboxyethyl)sulphanyl]-18-({2-[(3-{[4-({[({[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-1,2-dihydroxy-3,3-dimethylbutylidene]amino}-1-hydroxypropylidene)amino]ethyl}sulphanyl)-5-hydroxy-18-oxooctadeca-7,9,11,16-tetraenoic acid | HMDB |
|
|---|
| Chemical Formula | C42H61N8O22P3S2 |
|---|
| Average Molecular Mass | 1187.025 g/mol |
|---|
| Monoisotopic Mass | 1186.255 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 6-[(2-amino-2-carboxyethyl)sulfanyl]-18-{[2-(3-{3-[({[({[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido}propanamido)ethyl]sulfanyl}-5-hydroxy-18-oxooctadeca-7,9,11,16-tetraenoic acid |
|---|
| Traditional Name | 6-[(2-amino-2-carboxyethyl)sulfanyl]-18-({2-[3-(3-{[({[5-(6-aminopurin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-2-hydroxy-3-methylbutanamido)propanamido]ethyl}sulfanyl)-5-hydroxy-18-oxooctadeca-7,9,11,16-tetraenoic acid |
|---|
| SMILES | O[C@@H](CCCC(O)=O)[C@H](SC[C@@H](N)C(O)=O)\C=C\C=C\C=C/CCC\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP([O-])(=O)OP([O-])(=O)OC[C@H]1O[C@H]([C@@H](O)[C@H]1OP([O-])([O-])=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C42H65N8O22P3S2/c1-42(2,23-69-75(66,67)72-74(64,65)68-21-28-35(71-73(61,62)63)34(56)40(70-28)50-25-49-33-37(44)47-24-48-38(33)50)36(57)39(58)46-18-17-30(52)45-19-20-76-32(55)16-11-9-7-5-3-4-6-8-10-14-29(77-22-26(43)41(59)60)27(51)13-12-15-31(53)54/h3-4,6,8,10-11,14,16,24-29,34-36,40,51,56-57H,5,7,9,12-13,15,17-23,43H2,1-2H3,(H,45,52)(H,46,58)(H,53,54)(H,59,60)(H,64,65)(H,66,67)(H2,44,47,48)(H2,61,62,63)/p-4/b4-3-,8-6+,14-10+,16-11+/t26-,27+,28-,29-,34+,35+,36?,40-/m1/s1 |
|---|
| InChI Key | UNMYUSONOSUPAJ-WCKJHTGZSA-J |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as lipoxins. These are eicosanoids with a trihydroxyicosatetraenoic acid skeleton (a c20-fatty acid, with the chain bearing three hydroxyl groups and four double bonds). Lipoxins have four double bonds, which are all conjugated. In some cases a hydroxyl group is substituted by a C=O group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Eicosanoids |
|---|
| Direct Parent | Lipoxins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Lipoxin
- Long-chain fatty acid
- Hydroxy fatty acid
- Unsaturated fatty acid
- Fatty acid
- Enone
- Alpha,beta-unsaturated ketone
- Acryloyl-group
- 1,2-diol
- Secondary alcohol
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1900010000-3135169a1c613b6da357 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900223000-c34a5f83c51dba8d9b27 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900101000-e75816b073146289f0ae | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00e9-2900121200-22bdc63db637c299510e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-4900110000-aaa0bd962e1820bea153 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057r-6900100000-c489f195beab4c4e66fe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0019-4900000010-dcbd2ba333d072e0e7c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9100000001-51961232009f04fee79f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0036-9001200000-5ca6857a1f5d16c9e67a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-9400000003-a897c52f2667f469fbba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-6900000003-a40f2eb05858f79d65cc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-1210509000-4f438161d374db5c7e7f | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012597 |
|---|
| FooDB ID | FDB029140 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 76043766 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|