| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:05:14 UTC |
|---|
| Update Date | 2016-11-09 01:22:21 UTC |
|---|
| Accession Number | CHEM040937 |
|---|
| Identification |
|---|
| Common Name | 11R-HEPE |
|---|
| Class | Small Molecule |
|---|
| Description | An 11-HEPE that consists of (5Z,8Z,12E,14Z,17Z)-icosapentaenoic acid in which the hydroxy group is located at the 11R-position. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
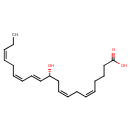
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (11R)-Hydroxy-(5Z,8Z,12E,14Z,17Z)-eicosapentaenoic acid | ChEBI | | (11R)-Hydroxy-(5Z,8Z,12E,14Z,17Z)-icosapentaenoic acid | ChEBI | | (5Z,8Z,11R,12E,14Z,17Z)-11-Hydroxyicosapentaenoic acid | ChEBI | | 11R-Hydroxy-5Z,8Z,12E,14Z,17Z-eicosapentaenoic acid | ChEBI | | (11R)-Hydroxy-(5Z,8Z,12E,14Z,17Z)-eicosapentaenoate | Generator | | (11R)-Hydroxy-(5Z,8Z,12E,14Z,17Z)-icosapentaenoate | Generator | | (5Z,8Z,11R,12E,14Z,17Z)-11-Hydroxyicosapentaenoate | Generator | | 11R-Hydroxy-5Z,8Z,12E,14Z,17Z-eicosapentaenoate | Generator | | 11(R)-Hydroxyeicosa-5Z,8Z,12E,14Z,17Z-pentaenoate | HMDB | | 11(R)-Hydroxyeicosa-5Z,8Z,12E,14Z,17Z-pentaenoic acid | HMDB |
|
|---|
| Chemical Formula | C20H30O3 |
|---|
| Average Molecular Mass | 318.450 g/mol |
|---|
| Monoisotopic Mass | 318.219 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (5Z,8Z,11R,12E,14Z,17Z)-11-hydroxyicosa-5,8,12,14,17-pentaenoic acid |
|---|
| Traditional Name | 11R-hepe |
|---|
| SMILES | CC\C=C/C\C=C/C=C/[C@H](O)C\C=C/C\C=C/CCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H30O3/c1-2-3-4-5-7-10-13-16-19(21)17-14-11-8-6-9-12-15-18-20(22)23/h3-4,6-7,9-11,13-14,16,19,21H,2,5,8,12,15,17-18H2,1H3,(H,22,23)/b4-3-,9-6-,10-7-,14-11-,16-13+/t19-/m0/s1 |
|---|
| InChI Key | IDEHSDHMEMMYIR-DJWFCICMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxyeicosapentaenoic acids. These are eicosanoic acids with an attached hydroxyl group and five CC double bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Eicosanoids |
|---|
| Direct Parent | Hydroxyeicosapentaenoic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydroxyeicosapentaenoic acid
- Long-chain fatty acid
- Hydroxy fatty acid
- Fatty acid
- Unsaturated fatty acid
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Organic oxygen compound
- Alcohol
- Hydrocarbon derivative
- Carbonyl group
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f6x-4952000000-cd3454cc64672f196c44 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00g1-9135200000-f174d03ce68e3fc5eb3f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0149000000-318dc0e1ccc495f069e8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0pi0-5973000000-8056f87480f9d53ad5d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9730000000-2c7ee4f4a41fe1a2d5bd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0059000000-92858f17a43771aacc45 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00r2-1293000000-c7d63c90b921bb316466 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abc-9730000000-060ffc1abb20c45f69ac | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0019000000-c475af61b990b381eca5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014j-0495000000-cb11bb0be17fb2f6002a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052e-9680000000-9f737f2d3c6c8180b634 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1549000000-1fc955f463e50f781964 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fza-2921000000-e098d6a463a3d5636949 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014l-9410000000-89939f3bb975a49a29fe | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012534 |
|---|
| FooDB ID | FDB029114 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4446310 |
|---|
| ChEBI ID | 91264 |
|---|
| PubChem Compound ID | 5283188 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|