| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:04:05 UTC |
|---|
| Update Date | 2016-11-09 01:22:21 UTC |
|---|
| Accession Number | CHEM040900 |
|---|
| Identification |
|---|
| Common Name | 7alpha-Hydroxy-3-oxo-4-cholestenoate |
|---|
| Class | Small Molecule |
|---|
| Description | A cholestanoid that is cholest-4-en-26-oic acid substituted by an alpha-hydroxy group at position 7 and an oxo group at position 3. It is an intermediate metabolite in the bile acid synthesis. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
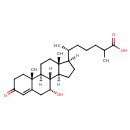
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 7-Hoca | ChEBI | | 7a-Hydroxy-3-oxo-4-cholestenoate | Generator | | 7a-Hydroxy-3-oxo-4-cholestenoic acid | Generator | | 7alpha-Hydroxy-3-oxo-4-cholestenoic acid | Generator | | 7Α-hydroxy-3-oxo-4-cholestenoate | Generator | | 7Α-hydroxy-3-oxo-4-cholestenoic acid | Generator | | 7 alpha-Hydroxy-3-oxo-4-cholestenoic acid | HMDB | | (7Α)-7-hydroxy-3-oxocholest-4-en-26-Oic acid | HMDB | | (7Α)-7-hydroxy-3-oxocholest-4-en-26-Oate | HMDB | | 7Α-hydroxy-3-oxocholest-4-en-26-Oic acid | HMDB | | (7alpha)-7-Hydroxy-3-oxocholest-4-en-26-Oic acid | HMDB | | (7alpha)-7-Hydroxy-3-oxocholest-4-en-26-Oate | HMDB | | 7alpha-Hydroxy-3-oxocholest-4-en-26-Oic acid | HMDB | | 7-alpha-Hydroxy-3-oxo-4-cholestenoate | HMDB | | 7alpha-Hydroxy-3-oxo-4-cholestenoate | HMDB, Generator |
|
|---|
| Chemical Formula | C27H42O4 |
|---|
| Average Molecular Mass | 430.620 g/mol |
|---|
| Monoisotopic Mass | 430.308 g/mol |
|---|
| CAS Registry Number | 115538-85-7 |
|---|
| IUPAC Name | (6R)-6-[(1S,2R,9R,10S,11S,14R,15R)-9-hydroxy-2,15-dimethyl-5-oxotetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-6-en-14-yl]-2-methylheptanoic acid |
|---|
| Traditional Name | (6R)-6-[(1S,2R,9R,10S,11S,14R,15R)-9-hydroxy-2,15-dimethyl-5-oxotetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-6-en-14-yl]-2-methylheptanoic acid |
|---|
| SMILES | C[C@H](CCCC(C)C(O)=O)C1CCC2C3[C@H](O)CC4=CC(=O)CC[C@]4(C)C3CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C27H42O4/c1-16(6-5-7-17(2)25(30)31)20-8-9-21-24-22(11-13-27(20,21)4)26(3)12-10-19(28)14-18(26)15-23(24)29/h14,16-17,20-24,29H,5-13,15H2,1-4H3,(H,30,31)/t16-,17?,20?,21?,22?,23-,24?,26+,27-/m1/s1 |
|---|
| InChI Key | SATGKQGFUDXGAX-KFNQEEOWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monohydroxy bile acids, alcohols and derivatives. These are bile acids, alcohols or any of their derivatives bearing a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Monohydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monohydroxy bile acid, alcohol, or derivatives
- Steroid acid
- 3-oxosteroid
- 3-oxo-delta-4-steroid
- 7-hydroxysteroid
- Oxosteroid
- Hydroxysteroid
- Delta-4-steroid
- Medium-chain fatty acid
- Cyclohexenone
- Hydroxy fatty acid
- Fatty acyl
- Cyclic alcohol
- Cyclic ketone
- Secondary alcohol
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organooxygen compound
- Organic oxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-0005900000-b7bca15cabd9e12c6696 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-029j-1009300000-55f0578d6b1582318cfc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-3219000000-f33373cace65dac79a13 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0001900000-ad4ad49ae3c3a393e0e8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-02dr-0007900000-88a170ea510b330ee78d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0avi-6009200000-78baf4ea8fd63c360662 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-2c11db7f4225ffd58c75 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0007900000-319ad83ee01108cd0a35 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-2003900000-183aa10ccc1b913cc96d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0004900000-6dd1885d6034e79dff91 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-3289300000-c62c18e4c41396c10579 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-059j-5971000000-425901eed2cf0114ca5a | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012458 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 2338765 |
|---|
| ChEBI ID | 83036 |
|---|
| PubChem Compound ID | 3081085 |
|---|
| Kegg Compound ID | C17337 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=17251592 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=19996111 | | 3. https://www.ncbi.nlm.nih.gov/pubmed/?term=8662693 | | 4. Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. | | 5. Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. | | 6. Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. | | 7. Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. | | 8. Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. | | 9. The lipid handbook with CD-ROM |
|
|---|