| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 01:03:56 UTC |
|---|
| Update Date | 2016-11-09 01:22:21 UTC |
|---|
| Accession Number | CHEM040897 |
|---|
| Identification |
|---|
| Common Name | 3alpha,7alpha,26-Trihydroxy-5beta-cholestane |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
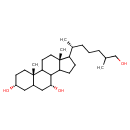
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3 a,7 a,26-Trihydroxy-5b-cholestane | Generator | | 3 Α,7 α,26-trihydroxy-5β-cholestane | Generator | | (25R)-5beta-Cholestane-3alpha,7alpha,26-triol | HMDB | | (3alpha,5beta,7alpha)-Cholestane-3,7,26-triol | HMDB | | 3alpha,7alpha,26-Trihydroxy-5beta-cholestane | HMDB | | 5 beta-Cholestane-3 alpha,7 alpha,26-triol | HMDB | | 5beta-Cholestan-3alpha,7alpha,26-triol | HMDB | | 5beta-Cholestane-3alpha,7alpha,26-triol | HMDB | | Cholestane-3,7,26-triol | HMDB |
|
|---|
| Chemical Formula | C27H48O3 |
|---|
| Average Molecular Mass | 420.668 g/mol |
|---|
| Monoisotopic Mass | 420.360 g/mol |
|---|
| CAS Registry Number | 15313-69-6 |
|---|
| IUPAC Name | (2S,5R,9R,15R)-14-[(2R)-7-hydroxy-6-methylheptan-2-yl]-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecane-5,9-diol |
|---|
| Traditional Name | (2S,5R,9R,15R)-14-[(2R)-7-hydroxy-6-methylheptan-2-yl]-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecane-5,9-diol |
|---|
| SMILES | CC(CO)CCC[C@@H](C)C1CCC2C3[C@H](O)CC4C[C@H](O)CC[C@]4(C)C3CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C27H48O3/c1-17(16-28)6-5-7-18(2)21-8-9-22-25-23(11-13-27(21,22)4)26(3)12-10-20(29)14-19(26)15-24(25)30/h17-25,28-30H,5-16H2,1-4H3/t17?,18-,19?,20-,21?,22?,23?,24-,25?,26+,27-/m1/s1 |
|---|
| InChI Key | OQIJRBFRXGIHMI-KZQGXEQDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Trihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - 26-hydroxysteroid
- Trihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 7-hydroxysteroid
- 3-alpha-hydroxysteroid
- Fatty alcohol
- Fatty acyl
- Cyclic alcohol
- Secondary alcohol
- Primary alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0bvi-0239300000-2d6fa65b7814b167dc14 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00di-1310149000-9437be2a819c430e4a74 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udr-0006900000-db130a37a7a56760612d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f79-1009300000-cb8caefba28321f5829d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05fr-2029000000-c0f180712508d632dc23 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0001900000-618009004df62f6c9bf9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uxr-0004900000-7a224517b07b79f9aa8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dr-2009100000-4d1414e27f71a6efe285 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uk9-0102900000-4b21863a4bcbc27f0eb3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0079-9124200000-cdd30e3905eb978af755 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-07kb-9420000000-0593538d66580bbdaca6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-0e0f641afc2bb1c607ec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0000900000-e524a811b2062225068c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-0002900000-2d299168fadc0554ef8f | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012455 |
|---|
| FooDB ID | FDB029071 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 28540 |
|---|
| PubChem Compound ID | 53481409 |
|---|
| Kegg Compound ID | C05444 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|