| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 00:32:05 UTC |
|---|
| Update Date | 2016-11-09 01:22:19 UTC |
|---|
| Accession Number | CHEM040771 |
|---|
| Identification |
|---|
| Common Name | Undecaprenyl N-acetyl-glucosaminyl-N-acetyl-mannosaminuronate-4-acetamido-4,6-dideoxy-D-galactose pyrophosphate |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Undecaprenyl N-acetyl-glucosaminyl-N-acetyl-mannosaminuronic acid-4-acetamido-4,6-dideoxy-D-galactose pyrophosphoric acid | Generator | | C55-PP-GlcNAc-mannaca-fuc4nac | HMDB | | Lipid III | HMDB | | ManNAcA-glcnac-fuc4nac-PP-lipid | HMDB | | ManNAcA-glcnac-fuc4nac-pyrophosphorylundecaprenol | HMDB | | Undecaprenyl-diphospho N-acetylglucosamine-N-acetylmannosaminuronate-N-acetamido-4,6-dideoxy-D-galactose | HMDB | | Ceramide nonasaccharide | MeSH, HMDB |
|
|---|
| Chemical Formula | C79H129N3O22P2 |
|---|
| Average Molecular Mass | 1534.824 g/mol |
|---|
| Monoisotopic Mass | 1533.854 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2S,3S,4R,5S,6R)-5-acetamido-3-{[(2R,3R,4S,6R)-5-acetamido-3,4-dihydroxy-6-methyloxan-2-yl]oxy}-6-{[(2R,3S,4R,5R,6R)-5-acetamido-4-hydroxy-6-{[hydroxy({[hydroxy({[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-2-(hydroxymethyl)oxan-3-yl]oxy}-4-hydroxyoxane-2-carboxylic acid |
|---|
| Traditional Name | (2S,3S,4R,5S,6R)-5-acetamido-3-{[(2R,3R,4S,6R)-5-acetamido-3,4-dihydroxy-6-methyloxan-2-yl]oxy}-6-{[(2R,3S,4R,5R,6R)-5-acetamido-4-hydroxy-6-{[hydroxy({hydroxy[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxyphosphoryl}oxy)phosphoryl]oxy}-2-(hydroxymethyl)oxan-3-yl]oxy}-4-hydroxyoxane-2-carboxylic acid |
|---|
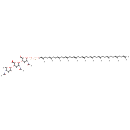
| SMILES | OC[C@H]1O[C@H](OP(=O)(O)OP(=O)(O)OC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CC\C=C(/C)CCC=C(C)C)[C@H](NC(C)=O)[C@@H](O)[C@@H]1O[C@@H]1O[C@H](C(=O)O)[C@@H](O[C@H]2O[C@H](C)C(NC(C)=O)[C@H](O)[C@H]2O)[C@H](O)[C@@H]1NC(=O)C |
|---|
| InChI Identifier | InChI=1S/C79H129N3O22P2/c1-50(2)27-17-28-51(3)29-18-30-52(4)31-19-32-53(5)33-20-34-54(6)35-21-36-55(7)37-22-38-56(8)39-23-40-57(9)41-24-42-58(10)43-25-44-59(11)45-26-46-60(12)47-48-97-105(93,94)104-106(95,96)103-78-68(82-64(16)86)70(88)73(65(49-83)99-78)100-77-67(81-63(15)85)71(89)74(75(102-77)76(91)92)101-79-72(90)69(87)66(61(13)98-79)80-62(14)84/h27,29,31,33,35,37,39,41,43,45,47,61,65-75,77-79,83,87-90H,17-26,28,30,32,34,36,38,40,42,44,46,48-49H2,1-16H3,(H,80,84)(H,81,85)(H,82,86)(H,91,92)(H,93,94)(H,95,96)/b51-29+,52-31+,53-33+,54-35+,55-37+,56-39+,57-41+,58-43+,59-45+,60-47+/t61-,65-,66?,67+,68-,69+,70-,71-,72-,73-,74+,75+,77-,78-,79-/m1/s1 |
|---|
| InChI Key | PSONHUYFSWYIME-JNYPHZTQSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as polyprenyl phospho carbohydrates. These are polyprenyl phosphates with a carbohydrate moiety attached to it. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Polyprenols |
|---|
| Direct Parent | Polyprenyl phospho carbohydrates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Polyterpenoid
- Bactoprenol diphosphate
- Polyprenyl phospho carbohydrate
- Oligosaccharide phosphate
- Polyprenyl monophosphate
- Oligosaccharide
- Polyprenyl phosphate skeleton
- N-acyl-alpha-hexosamine
- Glycosyl compound
- O-glycosyl compound
- Isoprenoid phosphate
- Organic pyrophosphate
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Alkyl phosphate
- Pyran
- Acetamide
- Secondary alcohol
- Carboxamide group
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Oxacycle
- Acetal
- Organopnictogen compound
- Alcohol
- Organic oxygen compound
- Organic nitrogen compound
- Primary alcohol
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Organooxygen compound
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0107651901-5d2fb76a53e28c193c83 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0101120900-d5a528f78dbfbd08a4e8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-0103220900-6240dba6699d3dfac8eb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004l-0642910002-2e026fe8d49f2e0d8162 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bt9-6855550709-3673ba5ef837620eac65 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-4910200003-281a64f8616d394629be | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0001090000-16fd8ce670774d49e820 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1193340000-9dbd03d5d4fc6fcffd74 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-4912050002-b947a659c9bd694b4e0b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053r-4000966103-ea9764d80655086db1a1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-3011964025-62a8e1cdbf10f9e88626 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0032-9212745800-09948d172da1ec184f3d | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012307 |
|---|
| FooDB ID | FDB028930 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35032433 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53481400 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|