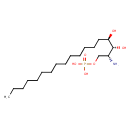| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 00:30:51 UTC |
|---|
| Update Date | 2016-11-09 01:22:19 UTC |
|---|
| Accession Number | CHEM040753 |
|---|
| Identification |
|---|
| Common Name | Phytosphingosine-1-phosphate |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| PHS-1-Phosphate | HMDB | | {[(2R,3R,4R)-2-amino-3,4-dihydroxyoctadecyl]oxy}phosphonate | Generator |
|
|---|
| Chemical Formula | C18H40NO6P |
|---|
| Average Molecular Mass | 397.487 g/mol |
|---|
| Monoisotopic Mass | 397.259 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | {[(2R,3R,4R)-2-amino-3,4-dihydroxyoctadecyl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3R,4R)-2-amino-3,4-dihydroxyoctadecyl]oxyphosphonic acid |
|---|
| SMILES | CCCCCCCCCCCCCC[C@@H](O)[C@H](O)[C@H](N)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H40NO6P/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-17(20)18(21)16(19)15-25-26(22,23)24/h16-18,20-21H,2-15,19H2,1H3,(H2,22,23,24)/t16-,17-,18-/m1/s1 |
|---|
| InChI Key | AYGOSKULTISFCW-KZNAEPCWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphosphingolipids. These are sphingolipids with a structure based on a sphingoid base that is attached to a phosphate head group. They differ from phosphonospingolipids which have a phosphonate head group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Phosphosphingolipids |
|---|
| Direct Parent | Phosphosphingolipids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sphingoid-1-phosphate or derivatives
- Phosphoethanolamine
- Monoalkyl phosphate
- Alkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- 1,3-aminoalcohol
- 1,2-diol
- 1,2-aminoalcohol
- Secondary alcohol
- Organopnictogen compound
- Organic nitrogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Primary aliphatic amine
- Amine
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01x0-2092000000-acc2f44cfa57dcd01b4c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-3009000000-8ebf59b673714024fe3b | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-3009000000-dfaf95a262f32fa73ae1 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-3009000000-b13c4300bd43cd6fe24d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udj-1109000000-ebc5dcf7ae08ad6f242e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f8c-6946000000-14b7651216a15069c22f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9100000000-793ba73f397ca7b12ee1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1009000000-6a89f2941d4431cc984a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9000000000-eed425d6e97a9cbe3c39 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012280 |
|---|
| FooDB ID | FDB031118 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | PHTYOSPHINGOSINE-1-P |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53481399 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|