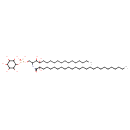| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 00:29:19 UTC |
|---|
| Update Date | 2016-11-09 01:22:19 UTC |
|---|
| Accession Number | CHEM040729 |
|---|
| Identification |
|---|
| Common Name | Inositol-P-ceramide |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| IPC | HMDB | | N-[3,4-Dihydroxy-1-({hydroxy[(2,3,4,5,6-pentahydroxycyclohexyl)oxy]phosphoryl}oxy)octadecan-2-yl]-2-hydroxyhexacosanimidate | Generator |
|
|---|
| Chemical Formula | C50H100NO13P |
|---|
| Average Molecular Mass | 954.302 g/mol |
|---|
| Monoisotopic Mass | 953.693 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | {[3,4-dihydroxy-2-(2-hydroxyhexacosanamido)octadecyl]oxy}[(2,3,4,5,6-pentahydroxycyclohexyl)oxy]phosphinic acid |
|---|
| Traditional Name | [3,4-dihydroxy-2-(2-hydroxyhexacosanamido)octadecyl]oxy(2,3,4,5,6-pentahydroxycyclohexyl)oxyphosphinic acid |
|---|
| SMILES | CCCCCCCCCCCCCCCCCCCCCCCCC(O)C(=O)NC(COP(O)(=O)OC1C(O)C(O)C(O)C(O)C1O)C(O)C(O)CCCCCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C50H100NO13P/c1-3-5-7-9-11-13-15-17-18-19-20-21-22-23-24-25-26-28-30-32-34-36-38-42(53)50(60)51-40(39-63-65(61,62)64-49-47(58)45(56)44(55)46(57)48(49)59)43(54)41(52)37-35-33-31-29-27-16-14-12-10-8-6-4-2/h40-49,52-59H,3-39H2,1-2H3,(H,51,60)(H,61,62) |
|---|
| InChI Key | PMXMKGYRVPAIJJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphosphingolipids. These are sphingolipids with a structure based on a sphingoid base that is attached to a phosphate head group. They differ from phosphonospingolipids which have a phosphonate head group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Phosphosphingolipids |
|---|
| Direct Parent | Phosphosphingolipids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sphingoid-1-phosphate or derivatives
- Inositol phosphate
- Phosphoethanolamine
- Cyclohexanol
- Dialkyl phosphate
- Cyclitol or derivatives
- Fatty amide
- Monosaccharide
- N-acyl-amine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Cyclic alcohol
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Carboxylic acid derivative
- Polyol
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Alcohol
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organic nitrogen compound
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f6x-5000003917-d10ceec7bb8592446bb6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00r7-9102002514-94d39f728e38ed187aad | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9101010110-804f0d2ec6bc7233337f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0000000009-71b1aa51b56e5ffae3a5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1021030019-41ae475e845a2fecf4f5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-9231000010-64118ccf5fdc255351ee | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0012237 |
|---|
| FooDB ID | FDB028880 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | IPC |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 24765754 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 25245972 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|