| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-27 00:19:11 UTC |
|---|
| Update Date | 2016-11-09 01:22:18 UTC |
|---|
| Accession Number | CHEM040662 |
|---|
| Identification |
|---|
| Common Name | (R)-2,3-Dihydroxy-3-methylvalerate |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
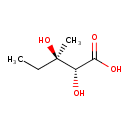
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2R,3R)-2,3-Dihydroxy-3-methylvaleric acid | ChEBI | | (2R,3R)-2,3-Dihydroxy-3-methylpentanoate | Kegg | | (2R,3R)-2,3-Dihydroxy-3-methylvalerate | Generator | | (2R,3R)-2,3-Dihydroxy-3-methylpentanoic acid | Generator | | (R)-2,3-Dihydroxy-3-methylvaleric acid | Generator | | (R) 2,3-Dihydroxy-3-methylvaleric acid | Generator, HMDB | | (R)-2,3-Dihydroxy-3-methylpentanoate | HMDB | | (R)-2,3-Dihydroxy-3-methylpentanoic acid | HMDB | | (R)-2,3-Dihydroxy-3-methylvalerate | HMDB, KEGG | | 1-keto-2-Methylvalerate | HMDB | | 2,3-Dihydroxy-3-methyl-valeric acid | HMDB | | 2,3-Dihydroxy-3-methylpentanoate | HMDB | | 2,3-Dihydroxy-3-methylpentanoic acid | HMDB | | 2,3-Dihydroxy-valerianic acid | HMDB | | 4,5-Dideoxy-3-C-methyl-pentonic acid | HMDB | | alpha,beta-Dihydroxy-beta-methylvaleric acid | HMDB | | CID8 | HMDB | | DMV | HMDB |
|
|---|
| Chemical Formula | C6H12O4 |
|---|
| Average Molecular Mass | 148.157 g/mol |
|---|
| Monoisotopic Mass | 148.074 g/mol |
|---|
| CAS Registry Number | 562-43-6 |
|---|
| IUPAC Name | (2R,3R)-2,3-dihydroxy-3-methylpentanoic acid |
|---|
| Traditional Name | (2R,3R)-2,3-dihydroxy-3-methylpentanoic acid |
|---|
| SMILES | CC[C@@](C)(O)[C@@H](O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H12O4/c1-3-6(2,10)4(7)5(8)9/h4,7,10H,3H2,1-2H3,(H,8,9)/t4-,6+/m0/s1 |
|---|
| InChI Key | PDGXJDXVGMHUIR-UJURSFKZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Hydroxy fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Branched fatty acid
- Methyl-branched fatty acid
- Short-chain hydroxy acid
- Hydroxy fatty acid
- Alpha-hydroxy acid
- Hydroxy acid
- Monosaccharide
- Tertiary alcohol
- 1,2-diol
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Alcohol
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00di-9000000000-d382043e81321879e92e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00tb-9464000000-0280de9eaa4c12333997 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05aj-7900000000-ed4255c918254a1dc462 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kar-9600000000-420faa83ea76001db2e9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pb9-9000000000-393b15b5811231e32693 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udj-2900000000-d582074beb8b89338fa5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9400000000-5ee3e425ebda8358d337 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05g0-9000000000-db72ead6bd87a129b325 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0h32-2900000000-0dc4c2a3c02313bed77c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-431db2e8dea59bdd5c38 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-349f8877717c1418480e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-81689c0553421abfcd88 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-d9815468ea9acf930e9e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9100000000-f894bf4ab44aaac79501 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB03675 |
|---|
| HMDB ID | HMDB0012140 |
|---|
| FooDB ID | FDB028798 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | 1-KETO-2-METHYLVALERATE |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | DMV |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 395044 |
|---|
| ChEBI ID | 27512 |
|---|
| PubChem Compound ID | 448154 |
|---|
| Kegg Compound ID | C06007 |
|---|
| YMDB ID | YMDB00068 |
|---|
| ECMDB ID | ECMDB12140 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|