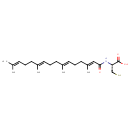| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 20:36:15 UTC |
|---|
| Update Date | 2016-11-09 01:22:15 UTC |
|---|
| Accession Number | CHEM040408 |
|---|
| Identification |
|---|
| Common Name | Geranylgeranylcysteine |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| S-(3,7,11,15-Tetramethyl-2,6,10,14-hexadecatetraenyl)-(e,e,e)-L-cysteine | HMDB | | S-[(2E,6E,10E)-3,7,11,15-Tetramethyl-2,6,10,14-hexadecatetraenyl]- (9ci)-L-cysteine | HMDB | | (2R)-2-{[(2E,6E,10E)-1-hydroxy-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-ylidene]amino}-3-sulfanylpropanoate | Generator, HMDB | | (2R)-2-{[(2E,6E,10E)-1-hydroxy-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-ylidene]amino}-3-sulphanylpropanoate | Generator, HMDB | | (2R)-2-{[(2E,6E,10E)-1-hydroxy-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-ylidene]amino}-3-sulphanylpropanoic acid | Generator, HMDB |
|
|---|
| Chemical Formula | C23H37NO3S |
|---|
| Average Molecular Mass | 407.610 g/mol |
|---|
| Monoisotopic Mass | 407.249 g/mol |
|---|
| CAS Registry Number | 169523-06-2 |
|---|
| IUPAC Name | (2R)-3-sulfanyl-2-[(2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenamido]propanoic acid |
|---|
| Traditional Name | (2R)-3-sulfanyl-2-[(2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenamido]propanoic acid |
|---|
| SMILES | SC[C@H](NC(=O)\C=C(/C)CC\C=C(/C)CC\C=C(/C)CCC=C(C)C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C23H37NO3S/c1-17(2)9-6-10-18(3)11-7-12-19(4)13-8-14-20(5)15-22(25)24-21(16-28)23(26)27/h9,11,13,15,21,28H,6-8,10,12,14,16H2,1-5H3,(H,24,25)(H,26,27)/b18-11+,19-13+,20-15+/t21-/m0/s1 |
|---|
| InChI Key | UBCKUGRWFMEXIF-WONWMLGISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyclic diterpenoids. These are diterpenoids (compounds made of four consecutive isoprene units) that do not contain a cycle. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Acyclic diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acyclic diterpenoid
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- N-acyl-l-alpha-amino acid
- Cysteine or derivatives
- Alpha-amino acid or derivatives
- N-acyl-amine
- Secondary carboxylic acid amide
- Carboxamide group
- Alkylthiol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Organosulfur compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000f-8889000000-3f81e03325547663218e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-022l-9352500000-a735063f6d3013ad3130 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-2769600000-ec3a2eaceb3ae67eb426 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ads-4961000000-44253e7d991961cef89d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ar0-8950000000-13cfd8c2ae6c7a125a61 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ab9-2017900000-84ecb7d4aa334cb77ecf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-059i-4329200000-97099be07e7e001431c7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-008l-9350000000-4a227b58aca7450e328b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-052r-0192200000-5cc299ff4ab00e2154d3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ei-2890000000-3dbfcbdc69c287f49fe6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00m0-7900000000-ebaf835d361f6930eb39 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0019000000-6bfbdf0cbf1824b2f5c6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-071l-9855100000-cb7896acb5225696b6a3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0536-9262000000-74db85fe9f128c11af64 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0011678 |
|---|
| FooDB ID | FDB028366 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 30776596 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53481025 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|