| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 20:34:26 UTC |
|---|
| Update Date | 2016-11-09 01:22:14 UTC |
|---|
| Accession Number | CHEM040358 |
|---|
| Identification |
|---|
| Common Name | Lactosyceramide (d18:1/18:1(9Z)) |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
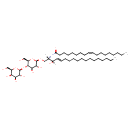
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| beta-D-Galactosyl-(1->4)-beta-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphing-4-enine | ChEBI | | beta-D-Galactosyl-(1->4)-beta-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphingosine | ChEBI | | beta-D-Galactosyl-(1->4)-beta-D-glucosyl-(11)-N-oleoylsphing-4-enine | ChEBI | | LacCer(D18:1/18:1(9Z)) | ChEBI | | N-(9Z-Octadecenoyl)-1-beta-lactosyl-sphing-4-enine | ChEBI | | N-(9Z-Octadecenoyl)-1-beta-lactosyl-sphingosine | ChEBI | | b-D-Galactosyl-(1->4)-b-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphing-4-enine | Generator | | Β-D-galactosyl-(1->4)-β-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphing-4-enine | Generator | | b-D-Galactosyl-(1->4)-b-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphingosine | Generator | | Β-D-galactosyl-(1->4)-β-D-glucosyl-(11)-N-[(9Z)-octadecenoyl]-sphingosine | Generator | | b-D-Galactosyl-(1->4)-b-D-glucosyl-(11)-N-oleoylsphing-4-enine | Generator | | Β-D-galactosyl-(1->4)-β-D-glucosyl-(11)-N-oleoylsphing-4-enine | Generator | | N-(9Z-Octadecenoyl)-1-b-lactosyl-sphing-4-enine | Generator | | N-(9Z-Octadecenoyl)-1-β-lactosyl-sphing-4-enine | Generator | | N-(9Z-Octadecenoyl)-1-b-lactosyl-sphingosine | Generator | | N-(9Z-Octadecenoyl)-1-β-lactosyl-sphingosine | Generator | | 1-O-(4-O-b-D-Galactopyranosyl-b-D-glucopyranosyl)-ceramide | HMDB | | 1-O-(4-O-beta-D-Galactopyranosyl-beta-glucopyranosyl)ceramide | HMDB | | 1-O-(4-O-beta-delta-Galactopyranosyl-beta-delta-glucopyranosyl)-ceramide | HMDB | | 1-O-(4-O-beta-delta-Galactopyranosyl-beta-glucopyranosyl)ceramide | HMDB | | 1Ylce-O-(4-O-beta-delta-galactopyranosyl-beta-glucopyranosyl)ceramide | HMDB | | beta-D-Galactosyl-1,4-beta-D-glucosylceramide | HMDB | | beta-delta-Galactosyl-1,4-beta-delta-glucosramide | HMDB | | beta-delta-Galactosyl-1,4-beta-delta-glucosylceramide | HMDB | | CDH | HMDB | | CDW17 Antigen | HMDB | | Cytolipin H | HMDB | | D-Galactosyl-1,4-beta-D-glucosylceramide | HMDB | | delta-Galactosyl-1,4-beta-delta-glucosylceramide | HMDB | | Gal-beta1->4GLC-beta1->1'cer | HMDB | | LacCer | HMDB | | LacCer(D18:1/18:1) | HMDB | | Lactosyl ceramide (D18:1/18:1(9Z)) | HMDB | | Lactosyl-N-acylsphingosine | HMDB | | Lactosylceramide | HMDB | | Lactosylceramide (D18:1/18:1) | HMDB | | N-(Oleoyl)-1-b-lactosyl-sphing-4-enine | HMDB | | N-(Oleoyl)-1-beta-lactosyl-sphing-4-enine | HMDB | | N-Lignoceryl sphingosyl lactoside | HMDB | | b-D-Galactosyl-(1->4)-b-D-glucosyl-(11)-N-oleoylsphingosine | HMDB | | Β-D-galactosyl-(1->4)-β-D-glucosyl-(11)-N-oleoylsphingosine | HMDB | | beta-D-Galactosyl-(1->4)-beta-D-glucosyl-(11)-N-oleoylsphingosine | HMDB |
|
|---|
| Chemical Formula | C48H89NO13 |
|---|
| Average Molecular Mass | 888.219 g/mol |
|---|
| Monoisotopic Mass | 887.633 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (9Z)-N-[(2S,3R,4E)-1-{[(2R,3R,4R,5S,6R)-3,4-dihydroxy-6-(hydroxymethyl)-5-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-2-yl]oxy}-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide |
|---|
| Traditional Name | (9Z)-N-[(2S,3R,4E)-1-{[(2R,3R,4R,5S,6R)-3,4-dihydroxy-6-(hydroxymethyl)-5-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-2-yl]oxy}-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide |
|---|
| SMILES | [H][C@@](CO[C@@H]1O[C@H](CO)[C@@H](O[C@@H]2O[C@H](CO)[C@H](O)[C@H](O)[C@H]2O)[C@H](O)[C@H]1O)(NC(=O)CCCCCCC\C=C/CCCCCCCC)[C@]([H])(O)\C=C\CCCCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C48H89NO13/c1-3-5-7-9-11-13-15-17-18-20-22-24-26-28-30-32-40(53)49-36(37(52)31-29-27-25-23-21-19-16-14-12-10-8-6-4-2)35-59-47-45(58)43(56)46(39(34-51)61-47)62-48-44(57)42(55)41(54)38(33-50)60-48/h17-18,29,31,36-39,41-48,50-52,54-58H,3-16,19-28,30,32-35H2,1-2H3,(H,49,53)/b18-17-,31-29+/t36-,37+,38+,39+,41-,42-,43+,44+,45+,46+,47+,48-/m0/s1 |
|---|
| InChI Key | YVBUQOZKCCPFCZ-HSVHIEHGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosyl-n-acylsphingosines. Glycosyl-N-acylsphingosines are compounds containing a sphingosine linked to a simple glucosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Glycosphingolipids |
|---|
| Direct Parent | Glycosyl-N-acylsphingosines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycosyl-n-acylsphingosine
- Fatty acyl glycoside
- Fatty acyl glycoside of mono- or disaccharide
- Alkyl glycoside
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Fatty amide
- N-acyl-amine
- Oxane
- Fatty acyl
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Organoheterocyclic compound
- Polyol
- Oxacycle
- Carboxylic acid derivative
- Acetal
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organonitrogen compound
- Carbonyl group
- Organooxygen compound
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0adi-0120033790-71f3ef58b41c7b419216 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0140251920-2bed52a9da05ddc7f02d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08g0-1690721530-823f8c3e54d8af0a9395 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00ri-0311031590-d6834701069a888036af | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0600-3633153940-fdece24178953f1ad714 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00c0-4920111200-defd723e60d70b12077f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0001000190-59c5ebc5001b558de665 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05mx-4592020320-51e23f5cafe9374e8e68 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ab9-6289030000-72efe06143bea41caa00 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01rb-2500090840-7d89f7559f706cda3c71 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-3800090310-3a152d728f4c4e9ceb67 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-090u-3900031000-d19a8ca95def6652c884 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0011592 |
|---|
| FooDB ID | FDB028305 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 24765751 |
|---|
| ChEBI ID | 131557 |
|---|
| PubChem Compound ID | 53480999 |
|---|
| Kegg Compound ID | C01290 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|