| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 19:26:48 UTC |
|---|
| Update Date | 2016-11-09 01:22:10 UTC |
|---|
| Accession Number | CHEM040001 |
|---|
| Identification |
|---|
| Common Name | L-Leucyl-L-proline |
|---|
| Class | Small Molecule |
|---|
| Description | A dipeptide formed from L-leucine and L-proline residues. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
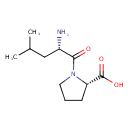
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Leu-L-pro | ChEBI | | L-P | ChEBI | | LP | ChEBI | | 1-L-Leucyl-L-proline | HMDB | | L-Leucyl-L-proline | HMDB | | L-p Dipeptide | HMDB | | LP Dipeptide | HMDB | | Leu-pro | HMDB | | Leucine proline dipeptide | HMDB | | Leucine-proline dipeptide | HMDB | | Leucyl-proline | HMDB | | Leucylproline | ChEBI |
|
|---|
| Chemical Formula | C11H20N2O3 |
|---|
| Average Molecular Mass | 228.288 g/mol |
|---|
| Monoisotopic Mass | 228.147 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2S)-1-[(2S)-2-amino-4-methylpentanoyl]pyrrolidine-2-carboxylic acid |
|---|
| Traditional Name | (2S)-1-[(2S)-2-amino-4-methylpentanoyl]pyrrolidine-2-carboxylic acid |
|---|
| SMILES | CC(C)CC(N)C(=O)N1CCCC1C(O)=O |
|---|
| InChI Identifier | InChI=1S/C11H20N2O3/c1-7(2)6-8(12)10(14)13-5-3-4-9(13)11(15)16/h7-9H,3-6,12H2,1-2H3,(H,15,16) |
|---|
| InChI Key | VTJUNIYRYIAIHF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Leucine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Proline or derivatives
- N-acyl-l-alpha-amino acid
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- N-acylpyrrolidine
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Pyrrolidine
- Tertiary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Azacycle
- Organoheterocyclic compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic oxide
- Organopnictogen compound
- Primary aliphatic amine
- Organic nitrogen compound
- Carbonyl group
- Amine
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01t9-1290000000-a7d4bfdd98e774a63ec6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02ti-9320000000-11c16428d6d853c82e2d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-066r-9000000000-1f0cc62f9745c1b92db3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-0690000000-abfdecce35ead141486f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01si-3930000000-da0f5e7072ac2161b71e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03mj-9600000000-7be81d5eb7b4a169cabc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0290000000-eeda503ca1f26574eff9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-5920000000-79943170b00abeee82ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03dj-9400000000-af008c1e3b0451414844 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-1290000000-a2306aff6fd871be8a62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-016s-9440000000-8aba88214d533f7870e0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9000000000-db271194855430c100f9 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0011175 |
|---|
| FooDB ID | FDB027947 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 72971 |
|---|
| ChEBI ID | 73580 |
|---|
| PubChem Compound ID | 80817 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Jandke J, Spiteller G: Dipeptide analysis in human urine. J Chromatogr. 1986 Oct 31;382:39-45. | | 2. Guo K, Li L: Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal Chem. 2009 May 15;81(10):3919-32. doi: 10.1021/ac900166a. | | 3. Lacha J, Hubacek JA, Potmesil P, Viklicky O, Malek I, Vitko S: TGF-beta I gene polymorphism in heart transplant recipients--effect on renal function. Ann Transplant. 2001;6(1):39-43. | | 4. Hebert EM, Mamone G, Picariello G, Raya RR, Savoy G, Ferranti P, Addeo F: Characterization of the pattern of alphas1- and beta-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. Appl Environ Microbiol. 2008 Jun;74(12):3682-9. doi: 10.1128/AEM.00247-08. Epub 2008 Apr 18. | | 5. Hebert EM, Raya RR, De Giori GS: Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. Appl Environ Microbiol. 2000 Dec;66(12):5316-21. | | 6. Liu X, Jin W, Theil EC: Opening protein pores with chaotropes enhances Fe reduction and chelation of Fe from the ferritin biomineral. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):3653-8. Epub 2003 Mar 12. | | 7. Cole A, Mendelblatt D, Aguayo J, Mathew A, Martin E, Vesely DL: Metastatic prostate cancer (with prostate-specific antigen of 9996) presenting as obstructive jaundice. Am J Med Sci. 2000 Feb;319(2):118-22. | | 8. Zhang F, Jia Z, Gao P, Kong H, Li X, Lu X, Wu Y, Xu G: Metabonomics study of urine and plasma in depression and excess fatigue rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Mol Biosyst. 2010 May;6(5):852-61. doi: 10.1039/b914751a. Epub 2010 Feb 9. | | 9. Martichonok V, Plouffe C, Storer AC, Menard R, Jones JB: Aziridine analogs of [[trans-(epoxysuccinyl)-L-leucyl]amino]-4-guanidinobutane (E-64) as inhibitors of cysteine proteases. J Med Chem. 1995 Aug 4;38(16):3078-85. | | 10. Guedon E, Renault P, Ehrlich SD, Delorme C: Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J Bacteriol. 2001 Jun;183(12):3614-22. | | 11. Marugg JD, Meijer W, van Kranenburg R, Laverman P, Bruinenberg PG, de Vos WM: Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol. 1995 Jun;177(11):2982-9. | | 12. Zhang F, Jia Z, Gao P, Kong H, Li X, Chen J, Yang Q, Yin P, Wang J, Lu X, Li F, Wu Y, Xu G: Metabonomics study of atherosclerosis rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Talanta. 2009 Aug 15;79(3):836-44. doi: 10.1016/j.talanta.2009.05.010. Epub 2009 May 19. | | 13. Pongracz E, Tordai A, Csornai M, Nagy Z: [Platelet glycoprotein IIb/IIIa (LeuPro 33) polymorphism in stroke patients]. Orv Hetil. 2001 Apr 15;142(15):781-5. | | 14. Osugi T, Ukena K, Bentley GE, O'Brien S, Moore IT, Wingfield JC, Tsutsui K: Gonadotropin-inhibitory hormone in Gambel's white-crowned sparrow (Zonotrichia leucophrys gambelii): cDNA identification, transcript localization and functional effects in laboratory and field experiments. J Endocrinol. 2004 Jul;182(1):33-42. | | 15. Huang Z, Lin L, Gao Y, Chen Y, Yan X, Xing J, Hang W: Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol Cell Proteomics. 2011 Oct;10(10):M111.007922. doi: 10.1074/mcp.M111.007922. Epub 2011 Jul 28. | | 16. Xu W, Chen D, Wang N, Zhang T, Zhou R, Huan T, Lu Y, Su X, Xie Q, Li L, Li L: Development of high-performance chemical isotope labeling LC-MS for profiling the human fecal metabolome. Anal Chem. 2015 Jan 20;87(2):829-36. doi: 10.1021/ac503619q. Epub 2014 Dec 25. |
|
|---|