| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 19:25:21 UTC |
|---|
| Update Date | 2016-11-09 01:22:10 UTC |
|---|
| Accession Number | CHEM039965 |
|---|
| Identification |
|---|
| Common Name | LysoPE(18:0/0:0) |
|---|
| Class | Small Molecule |
|---|
| Description | A 1-acyl-sn-glycero-3-phosphoethanolamine zwitterion obtained by transfer of a proton from the amino to the phosphate group of 1-stearoyl-sn-glycero-3-phosphoethanolamine. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
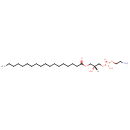
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-C18:0-Phosphatidylethanolamine zwitterion | ChEBI | | 1-Octadecanoyl-sn-glycero-3-phosphoethanolamine | ChEBI | | 1-Octadecanoyl-sn-glycero-3-phosphoethanolamine zwitterion | ChEBI | | 2-Azaniumylethyl (2R)-2-hydroxy-3-(stearoyloxy)propyl phosphate | ChEBI | | 2-Azaniumylethyl (2R)-2-hydroxy-3-(stearoyloxy)propyl phosphoric acid | Generator | | octadecanoyl-lysophosphatidylethanolamine | Lipid Annotator, HMDB | | 1-stearoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | Lipid Annotator, HMDB | | LPE(18:0) | Lipid Annotator, HMDB | | LPE(18:0/0:0) | Lipid Annotator, HMDB | | Lysophosphatidylethanolamine(18:0/0:0) | Lipid Annotator, HMDB | | Lysophosphatidylethanolamine(18:0) | Lipid Annotator, HMDB | | Lyso-PE(18:0/0:0) | Lipid Annotator, HMDB | | Lyso-PE(18:0) | Lipid Annotator, HMDB | | LysoPE(18:0/0:0) | Lipid Annotator | | 1-octadecanoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | Lipid Annotator, HMDB | | LysoPE(18:0) | Lipid Annotator, HMDB | | Stearoyl phosphatidylethanolamine | HMDB | | 1-Octadecanoylglycerophosphoethanolamine | HMDB | | 1-Octadecanoyl-2-hydroxy-sn-glycerol-3-phosphatidyl ethanolamine | HMDB | | 1-Stearoyl-3-glycerylphosphorylethanolamine | HMDB | | 1-Stearoylglycerophosphoethanolamine | HMDB | | Stearoyl lysophophatidylethanolamine | HMDB | | 1-Stearoyl-GPE | HMDB | | 1-Stearoyl-lysophosphatidylethanolamine | HMDB | | 1-Stearoyl-sn-glycero-3-phosphoethanolamine | HMDB | | GPE(18:0) | HMDB | | GPE(18:0/0:0) | HMDB |
|
|---|
| Chemical Formula | C23H48NO7P |
|---|
| Average Molecular Mass | 481.604 g/mol |
|---|
| Monoisotopic Mass | 481.317 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (2-aminoethoxy)[(2R)-2-hydroxy-3-(octadecanoyloxy)propoxy]phosphinic acid |
|---|
| Traditional Name | 2-aminoethoxy(2R)-2-hydroxy-3-(octadecanoyloxy)propoxyphosphinic acid |
|---|
| SMILES | [H][C@@](O)(COC(=O)CCCCCCCCCCCCCCCCC)COP(O)(=O)OCCN |
|---|
| InChI Identifier | InChI=1S/C23H48NO7P/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-23(26)29-20-22(25)21-31-32(27,28)30-19-18-24/h22,25H,2-21,24H2,1H3,(H,27,28)/t22-/m1/s1 |
|---|
| InChI Key | BBYWOYAFBUOUFP-JOCHJYFZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-acyl-sn-glycero-3-phosphoethanolamines. These are glycerophoethanolamines in which the glycerol is esterified with a fatty acid at O-1 position, and linked at position 3 to a phosphoethanolamine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoethanolamines |
|---|
| Direct Parent | 1-acyl-sn-glycero-3-phosphoethanolamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-monoacyl-sn-glycero-3-phosphoethanolamine
- Phosphoethanolamine
- Fatty acid ester
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Amino acid or derivatives
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organonitrogen compound
- Hydrocarbon derivative
- Alcohol
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organooxygen compound
- Primary amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udu-3940200000-708255d7f203656bccb8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0fa6-9650030000-578345aff8fc04704aa2 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9121200000-0cc3db284eda8ba9bf93 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9121000000-c3045d676cec69c82c2c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9120000000-c6389552dc63301a44b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00lr-1290500000-5cb171232cddea4bcd41 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00o3-6490100000-990e3741062f22dda9f8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-9110000000-b787134651d069f7710e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-3003900000-80b41628e9438152c12f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9104200000-dbc3c048c121ba2954d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9300000000-7c39510d12d6cdac8016 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000900000-270fd00e9abe947aaec1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1390400000-cfd942fee43ce80548cc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1390000000-0af873e0b2e5ea7259d2 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0011130 |
|---|
| FooDB ID | FDB027909 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 7826018 |
|---|
| ChEBI ID | 75036 |
|---|
| PubChem Compound ID | 9547068 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | YMDB01204 |
|---|
| ECMDB ID | M2MDB003803 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Divecha N, Irvine RF: Phospholipid signaling. Cell. 1995 Jan 27;80(2):269-78. | | 2. Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. | | 3. Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. | | 4. Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. | | 5. Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. | | 6. Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. | | 7. Phospholipids Handbook | | 8. The lipid handbook with CD-ROM |
|
|---|