| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:39:07 UTC |
|---|
| Update Date | 2016-11-09 01:21:17 UTC |
|---|
| Accession Number | CHEM035307 |
|---|
| Identification |
|---|
| Common Name | 8-Hydroxyguanine |
|---|
| Class | Small Molecule |
|---|
| Description | An oxopurine that is guanine in which the hydrogen at position 8 is replaced by an oxo group and in which the nitrogens at positions 7 and 9 each bear a hydrogen. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
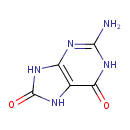
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 8-Oxoguanine | ChEBI | | 1H-Purine-6,8-dione, 2-amino-7,9-dihydro-1H-purine-6,8-dione | HMDB | | 2-amino-6,8-Dihydroxypurine | HMDB, MeSH | | 2-amino-7,9-dihydro-1H-Purine-6,8-dione | HMDB | | 2-amino-Purine-6,8-diol | HMDB | | 2-Aminopurine-6,8-diol | HMDB | | 7,8-dihydro-8-Oxoguanine | HMDB | | oxo-8-Gua | MeSH, HMDB | | 8-oxo-7,8-Dihydroguanine | MeSH, HMDB | | 8-Hydroxyguanine | ChEBI |
|
|---|
| Chemical Formula | C5H5N5O2 |
|---|
| Average Molecular Mass | 167.126 g/mol |
|---|
| Monoisotopic Mass | 167.044 g/mol |
|---|
| CAS Registry Number | 5614-64-2 |
|---|
| IUPAC Name | 2-amino-6,7,8,9-tetrahydro-1H-purine-6,8-dione |
|---|
| Traditional Name | 8-hydroxyguanine |
|---|
| SMILES | NC1=NC2=C(NC(=O)N2)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C5H5N5O2/c6-4-8-2-1(3(11)10-4)7-5(12)9-2/h(H5,6,7,8,9,10,11,12) |
|---|
| InChI Key | CLGFIVUFZRGQRP-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purines and purine derivatives. These are aromatic heterocyclic compounds containing a purine moiety, which is formed a pyrimidine-ring ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Purines and purine derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine
- Hydroxypyrimidine
- Pyrimidine
- Heteroaromatic compound
- Imidazole
- Azole
- Azacycle
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-014i-3900000000-58549cee828db799d7b5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-6a4117b10e5530095707 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-89fdc38268686eaba66a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-107l-9600000000-7c79695abe7a15874725 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0900000000-ac22897692beb4981ede | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-2900000000-f3b3767a022d716580d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-9a8d26eff2e742411cb2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0900000000-d3890f3472d103400e66 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00ri-1900000000-59a4be35534659234b6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-7d9830f9bdf455a84283 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-2907e7cb009d3747cf21 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-5795d2965bc15c5daf00 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00us-9200000000-fd1acc84bba1739821db | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0002032 |
|---|
| FooDB ID | FDB022809 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | 8-Oxoguanine |
|---|
| Chemspider ID | 58661 |
|---|
| ChEBI ID | 52617 |
|---|
| PubChem Compound ID | 65154 |
|---|
| Kegg Compound ID | C20155 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|