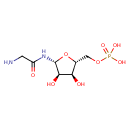| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:38:56 UTC |
|---|
| Update Date | 2016-11-09 01:21:17 UTC |
|---|
| Accession Number | CHEM035303 |
|---|
| Identification |
|---|
| Common Name | Glycineamideribotide |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GAR | Kegg | | N1-(5-Phospho-D-ribosyl)glycinamide | Kegg | | Glycinamide ribonucleotide | Kegg | | N1-(5-Phospho-beta-D-ribosyl)glycinamide | Kegg | | N1-(5-Phospho-b-D-ribosyl)glycinamide | Generator | | N1-(5-Phospho-β-D-ribosyl)glycinamide | Generator | | 5'-Phosphoribosyl-glycineamide | HMDB | | 5'-Phosphoribosylglycinamide | HMDB | | 5'-Phosphoribosylglycineamide | HMDB | | Glycineamide ribonucleotide | HMDB | | N(1)-(5-phospho-D-Ribosyl)glycinamide | HMDB | | N-Glycyl-5-O-phosphono-D-ribofuranosylamine | HMDB | | 2-Amino-N-(5-O-phosphono-beta-D-ribofuranosyl)acetamide | HMDB | | 2-Amino-N-(5-O-phosphono-β-D-ribofuranosyl)acetamide | HMDB | | 5-Phospho-beta-D-ribosyl-glycineamide | HMDB | | 5-Phospho-β-D-ribosyl-glycineamide | HMDB | | 5’-Phosphoribosylglycinamide | HMDB | | 5’-Phosphoribosylglycineamide | HMDB |
|
|---|
| Chemical Formula | C7H15N2O8P |
|---|
| Average Molecular Mass | 286.176 g/mol |
|---|
| Monoisotopic Mass | 286.057 g/mol |
|---|
| CAS Registry Number | 10074-18-7 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(2-aminoacetamido)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | glycineamide ribonucleotide |
|---|
| SMILES | NCC(=O)N[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C7H15N2O8P/c8-1-4(10)9-7-6(12)5(11)3(17-7)2-16-18(13,14)15/h3,5-7,11-12H,1-2,8H2,(H,9,10)(H2,13,14,15)/t3-,5-,6-,7-/m1/s1 |
|---|
| InChI Key | OBQMLSFOUZUIOB-SHUUEZRQSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycinamide ribonucleotides. Glycinamide ribonucleotides are compounds in which the amide N atom of glycineamide is linked to the C-1 of a ribosyl (or deoxyribosyl) moiety. Nucleotides have a phosphate group linked to the C5 carbon of the ribose (or deoxyribose) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Glycinamide ribonucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Glycinamide ribonucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycinamide-ribonucleotide
- Pentose phosphate
- Pentose-5-phosphate
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- Monosaccharide phosphate
- Pentose monosaccharide
- Monoalkyl phosphate
- Monosaccharide
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Tetrahydrofuran
- Secondary carboxylic acid amide
- Secondary alcohol
- Amino acid or derivatives
- Carboxamide group
- 1,2-diol
- Organoheterocyclic compound
- Oxacycle
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxide
- Primary aliphatic amine
- Organopnictogen compound
- Alcohol
- Organonitrogen compound
- Carbonyl group
- Organooxygen compound
- Amine
- Organic oxygen compound
- Hydrocarbon derivative
- Primary amine
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000t-9420000000-d7fee78dd4657e2eba28 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00si-9541200000-4a23e3091baf72b01dc2 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053i-9460000000-f71dc4d8ec0f4da03819 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0560-9120000000-4770933b45387be6728c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9200000000-a6f2b9ab201fd0e69ee8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0170-9550000000-99520656892b8c415efb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-d6c860661b01956c80ac | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-c2f2b85e6ec9caaa4408 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-2090000000-f6fcd99577c7e5d5d2ec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9020000000-f5f361d4aef59382fc15 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-962cd48ee45a1c95e446 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0980000000-6a9e014a86b77c5ad3c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-5920000000-cb0c5e3520ad275d3742 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-057j-9500000000-ab2ea8c5eb8a25044810 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0002022 |
|---|
| FooDB ID | FDB022801 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00007395 |
|---|
| BiGG ID | 42621 |
|---|
| BioCyc ID | 5-PHOSPHO-RIBOSYL-GLYCINEAMIDE |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Glycineamide ribonucleotide |
|---|
| Chemspider ID | 141370 |
|---|
| ChEBI ID | 18349 |
|---|
| PubChem Compound ID | 160913 |
|---|
| Kegg Compound ID | C03838 |
|---|
| YMDB ID | YMDB00374 |
|---|
| ECMDB ID | ECMDB01567 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|