| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:38:09 UTC |
|---|
| Update Date | 2016-11-09 01:21:17 UTC |
|---|
| Accession Number | CHEM035288 |
|---|
| Identification |
|---|
| Common Name | 1,7-Dimethylguanosine |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
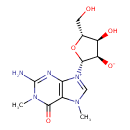
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,7-Dimethylguanosine | MeSH |
|
|---|
| Chemical Formula | C12H17N5O5 |
|---|
| Average Molecular Mass | 311.294 g/mol |
|---|
| Monoisotopic Mass | 311.123 g/mol |
|---|
| CAS Registry Number | 69453-64-1 |
|---|
| IUPAC Name | 2-amino-9-[(2R,3R,4S,5R)-4-hydroxy-5-(hydroxymethyl)-3-oxidooxolan-2-yl]-1,7-dimethyl-6-oxo-6,7-dihydro-1H-9λ⁵-purin-9-ylium |
|---|
| Traditional Name | 2-amino-9-[(2R,3R,4S,5R)-4-hydroxy-5-(hydroxymethyl)-3-oxidooxolan-2-yl]-1,7-dimethyl-6-oxo-9λ⁵-purin-9-ylium |
|---|
| SMILES | CN1C=[N+]([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2[O-])C2=C1C(=O)N(C)C(N)=N2 |
|---|
| InChI Identifier | InChI=1S/C12H17N5O5/c1-15-4-17(11-8(20)7(19)5(3-18)22-11)9-6(15)10(21)16(2)12(13)14-9/h4-5,7-8,11,18-19H,3H2,1-2H3,(H2,13,14)/t5-,7-,8-,11-/m1/s1 |
|---|
| InChI Key | AISVAXNALHBFPC-IOSLPCCCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Purine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleoside
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Pyrimidone
- Monosaccharide
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Secondary alcohol
- Lactam
- Organoheterocyclic compound
- Oxacycle
- Azacycle
- Primary amine
- Organonitrogen compound
- Organooxygen compound
- Primary alcohol
- Amine
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Organic zwitterion
- Alkoxide
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ul3-5190000000-b6efd1b0de6f867dfef1 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0pdi-6914300000-55a8e845530a17b1def6 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0901000000-ff3d53947e369807a1a6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-644d98aff8beb3c230d8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-0900000000-ed639b094f6f657cdd6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03e9-0978000000-d999459b8228690b0347 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0920000000-7f9f2e8bd0a05e57db97 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03di-2900000000-dae0f412572a1d88a824 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0902000000-92506f2955e30691cc39 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-bf2000feeb7b932cc0b1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-2910000000-0f7615343b7eb1969bfa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-0911000000-5748212a364a9de4d004 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0900000000-e17f62224c72bd0c388a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00fr-2910000000-70e877dfb2209a63c9c1 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001961 |
|---|
| FooDB ID | FDB022767 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 140757 |
|---|
| ChEBI ID | 165836 |
|---|
| PubChem Compound ID | 160130 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|