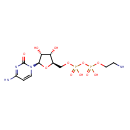
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004i-6793400000-ac76da0c6087752db3c9 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00pj-8972160000-fd358ed61430e64ed4e0 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 17V, negative | splash10-0002-0000900000-0b86b2b2cfc17d95ff90 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 19V, negative | splash10-0002-1012900000-c5804a13d813a5377b79 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 23V, negative | splash10-0032-8157900000-36b36bcbce04a95bf962 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 29V, negative | splash10-004i-9122000000-f9d8d35bd43030c78e8b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 35V, negative | splash10-004i-9100000000-8db373393648433147a0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 42V, negative | splash10-004i-9100000000-1cb47f991b97749fba0b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 50V, negative | splash10-004i-9000000000-4a5fcad27ccf2ff23957 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 63V, negative | splash10-004i-9000000000-6b08787f67155728185f | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 31V, negative | splash10-001i-0029000000-1668461b10d942d91cf9 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 31V, negative | splash10-004i-9300000000-8eef8ed8fdaa72ae58a8 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 31V, negative | splash10-03fr-1390000000-88ee14c068713b4ad543 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 31V, negative | splash10-00di-0290000000-3006b045a5e65ff5fa52 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 31V, negative | splash10-0a4i-0900000000-4a998d56bcc8853cc4ae | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 53V, negative | splash10-0002-3012900000-be071eccb64015240644 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 64V, negative | splash10-004i-9124300000-5d5dc9798e172dc0a1e0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 78V, negative | splash10-004i-9100000000-c769c60b7993983641f0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 92V, negative | splash10-004i-9100000000-1837fdf24962abbe6805 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, negative | splash10-001i-0009000000-0d7a52eebf0db4a36d24 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 20V, negative | splash10-001i-2129000000-a4a5a49c30ed6207c333 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-5910100000-2a4aae3a59f3f928bc14 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-7900000000-70f4dc43398ae42bd718 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9700000000-2d6893ab115d449880c3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gvk-1512900000-bd91f3bd3454ae8dc4da | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0n4i-6942200000-8c75516485feb49d2fa1 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tf-5900000000-624299483014ca3a86f4 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |