| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:35:33 UTC |
|---|
| Update Date | 2016-11-09 01:21:16 UTC |
|---|
| Accession Number | CHEM035228 |
|---|
| Identification |
|---|
| Common Name | Guanosine triphosphate adenosine |
|---|
| Class | Small Molecule |
|---|
| Description | Guanosine triphosphate adenosine is a dinucleoside polyphosphate. Dinucleoside polyphosphates are an interesting group of signalling molecules that control numerous physiological functions. Diadenosine compounds, with a backbone of anything from two to seven phosphates, are known to occur naturally. Some of them have been isolated from cerebral nerve terminals and, acting via nucleoside (P1), nucleotide (P2), or dinucleotide receptors, can affect central nervous system function. Many of them have been isolated from human blood platelet secretory granules and are potentially involved in haemostatic mechanisms and peripheral control of vascular tone. Many visceral organs respond to the application of adenine dinucleotides and, although they act on receptors in the periphery that can be mainly defined as either P1 or P2, evidence is now accumulating for discrete dinucleotide receptors. In the periphery, adenine dinucleotides can be potent agonists, with diverse functions, causing contraction or relaxation of smooth muscle. Many P2X receptor proteins and P2Y receptors have been cloned and adenine dinucleotides have a variable pharmacological profile at these receptors and may be useful tools for characterising subtypes of P2X and P2Y receptors. Many extracellular roles of diadenosine polyphosphates are emerging as yet increasingly important, natural ligands for a plethora of structurally diverse mononucleotide and dinucleotide receptors. (PMID: 12772275, 7767329). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
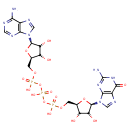
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Guanosine triphosphoric acid adenosine | Generator | | Guanosine 5'-triphosphate-5'-adenosine | HMDB | | Guanosine triphosphate, 5'>5'-ester with adenosine (7ci) | HMDB | | P1-Adenosine-5' P3-guanosine-5' triphosphate | HMDB | | [({[(2R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][({[(2R,5R)-3,4-dihydroxy-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphinate | Generator, HMDB |
|
|---|
| Chemical Formula | C20H27N10O17P3 |
|---|
| Average Molecular Mass | 772.407 g/mol |
|---|
| Monoisotopic Mass | 772.077 g/mol |
|---|
| CAS Registry Number | 10527-47-6 |
|---|
| IUPAC Name | {[(2R,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({[({[(2R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphinic acid |
|---|
| Traditional Name | [(2R,5R)-5-(2-amino-6-oxo-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy({[(2R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxyphosphinic acid |
|---|
| SMILES | NC1=NC2=C(N=CN2[C@@H]2O[C@H](COP(O)(=O)OP(O)(=O)OP(O)(=O)OC[C@H]3O[C@H](C(O)C3O)N3C=NC4=C3N=CN=C4N)C(O)C2O)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C20H27N10O17P3/c21-14-8-15(24-3-23-14)29(4-25-8)18-12(33)10(31)6(44-18)1-42-48(36,37)46-50(40,41)47-49(38,39)43-2-7-11(32)13(34)19(45-7)30-5-26-9-16(30)27-20(22)28-17(9)35/h3-7,10-13,18-19,31-34H,1-2H2,(H,36,37)(H,38,39)(H,40,41)(H2,21,23,24)(H3,22,27,28,35)/t6-,7-,10?,11?,12?,13?,18-,19-/m1/s1 |
|---|
| InChI Key | PMJUJCUPNRCBNP-KNKPYYCGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as (5'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (5'->5')-phosphodiester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | (5'->5')-dinucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | (5'->5')-dinucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - (5'->5')-dinucleotide
- Purine ribonucleoside triphosphate
- Purine nucleotide sugar
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Imidazopyrimidine
- Purine
- Pyrimidone
- Monoalkyl phosphate
- Aminopyrimidine
- Pyrimidine
- Alkyl phosphate
- Imidolactam
- Phosphoric acid ester
- Monosaccharide
- Organic phosphoric acid derivative
- N-substituted imidazole
- Tetrahydrofuran
- Vinylogous amide
- Azole
- Imidazole
- Heteroaromatic compound
- Secondary alcohol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Amine
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05i9-0212190200-bdb4b048673947df6734 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_10) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-0910000100-0542cf2c5f05bccb6cdf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f79-0900000000-7280c02dd129d69e7668 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f79-0900000000-8e745cc2f83b67dc2d01 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ul0-0900020500-6147fda0a5749a549d11 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f89-1900000000-8f6c343a5efb9242e224 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0kar-1940000000-d9377b149e23683b439f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-d23f2a6416fd8d87a443 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udr-0900000100-6c8bbc4df2b5fa2e8904 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f79-0910000000-e95c8809be84422e4a5c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000000900-a1ec46398398344054a9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ff0-1900012600-a289155ace6733ef1e0a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0610900200-6834319ffc1b39f4854e | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001459 |
|---|
| FooDB ID | FDB022637 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013035 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 53477730 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|