| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:34:29 UTC |
|---|
| Update Date | 2016-11-09 01:21:16 UTC |
|---|
| Accession Number | CHEM035205 |
|---|
| Identification |
|---|
| Common Name | 3-Sulfinylpyruvic acid |
|---|
| Class | Small Molecule |
|---|
| Description | 3-Sulfinylpyruvic acid, also known as 3-sulfinylpyruvic acid or 3-sulfinylpyruvic acid, belongs to the class of organic compounds known as alpha-keto acids and derivatives. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. 3-Sulfinylpyruvic acid is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). 3-Sulfinylpyruvic acid exists in all living organisms, ranging from bacteria to humans. 3-Sulfinylpyruvic acid and L-glutamic acid can be biosynthesized from 3-sulfinoalanine and oxoglutaric acid; which is catalyzed by the enzyme aspartate aminotransferase, cytoplasmic. In cattle, 3-sulfinylpyruvic acid is involved in the metabolic pathway called the cysteine metabolism pathway. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
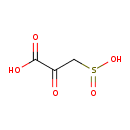
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Sulfinopyruvate | ChEBI | | 3-Sulfinylpyruvate | ChEBI | | 3-Sulfinopyruvic acid | Generator | | 3-Sulphinopyruvate | Generator | | 3-Sulphinopyruvic acid | Generator | | 3-Sulphinylpyruvate | Generator | | 3-Sulphinylpyruvic acid | Generator | | beta-Sulfinyl pyruvate | HMDB | | beta-Sulfinyl pyruvic acid | HMDB | | 2-oxo-3-Sulfinopropanoic acid | HMDB | | beta-Sulfinyl-pyruvate | HMDB | | beta-Sulfinyl-pyruvic acid | HMDB | | Β-sulfinyl pyruvate | HMDB | | Β-sulfinyl pyruvic acid | HMDB | | Β-sulfinyl-pyruvate | HMDB | | Β-sulfinyl-pyruvic acid | HMDB | | 3-Sulfinylpyruvic acid | Generator |
|
|---|
| Chemical Formula | C3H3O5S |
|---|
| Average Molecular Mass | 151.118 g/mol |
|---|
| Monoisotopic Mass | 150.970 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 2-oxo-3-sulfinopropanoic acid |
|---|
| Traditional Name | 3-sulfinylpyruvate |
|---|
| SMILES | OC(=O)C(=O)CS([O-])=O |
|---|
| InChI Identifier | InChI=1S/C3H4O5S/c4-2(3(5)6)1-9(7)8/h1H2,(H,5,6)(H,7,8)/p-1 |
|---|
| InChI Key | JXYLQEMXCAAMOL-UHFFFAOYSA-M |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha-keto acids and derivatives. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Alpha-keto acids and derivatives |
|---|
| Direct Parent | Alpha-keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-keto acid
- Alpha-hydroxy ketone
- Sulfinic acid
- Ketone
- Alkanesulfinic acid
- Alkanesulfinic acid or derivatives
- Sulfinic acid derivative
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxygen compound
- Carbonyl group
- Organic oxide
- Organooxygen compound
- Organosulfur compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0bvl-9300000000-b9d5aeadd8b7738b7353 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0ab9-9600000000-56bba3bcd18999fead91 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-0900000000-871c9b0796f2447470ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00li-9600000000-da362e24b25de10e7c3e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-47872b5d21e6463c1ffc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-1900000000-d91ad6a6d2809d9a6714 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fb9-9400000000-f213ed656a1bcd5e1554 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0w93-9400000000-7973a19a971bd544bf7e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ulc-9600000000-23622faf57828eab4fed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-9000000000-4fae7c347514ef087b68 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9000000000-5f2edaa9051f5ab324ba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-9216d677abe986964826 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-9000000000-64a44662186d1bb88d15 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-16b11026ba456f1a6f3c | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001405 |
|---|
| FooDB ID | FDB022965 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | 6627 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 108 |
|---|
| ChEBI ID | 1665 |
|---|
| PubChem Compound ID | 110 |
|---|
| Kegg Compound ID | C05527 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | ECMDB01405 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|