| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:29:21 UTC |
|---|
| Update Date | 2016-11-09 01:21:15 UTC |
|---|
| Accession Number | CHEM035099 |
|---|
| Identification |
|---|
| Common Name | 3-Hydroxybutyryl-CoA |
|---|
| Class | Small Molecule |
|---|
| Description | 3-Hydroxybutyryl-CoA is a substrate for Enoyl-CoA hydratase (mitochondrial), Trifunctional enzyme alpha subunit (mitochondrial) and Peroxisomal bifunctional enzyme. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
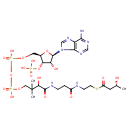
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3R)-3-Hydroxybutanoyl-CoA | HMDB | | (3R)-3-Hydroxybutanoyl-coenzyme A | HMDB | | (R)-3-Hydroxybutanoyl-CoA | HMDB | | (R)-3-Hydroxybutanoyl-coenzyme A | HMDB | | (S)-3-Hydroxybutanoyl-CoA | HMDB | | (S)-3-Hydroxybutanoyl-coenzyme A | HMDB | | 3-Hydroxybutanoyl-CoA | HMDB | | 3-Hydroxybutanoyl-coenzyme A | HMDB | | 3-Hydroxybutyryl-coenzyme A | HMDB | | 3-OH-Butyryl-CoA | HMDB | | 3-OH-Butyryl-coenzyme A | HMDB | | beta-Hydroxybutyryl-CoA | HMDB | | beta-Hydroxybutyryl-coenzyme A | HMDB | | beta-Hydroxybutyryl-S-CoA | HMDB | | beta-Hydroxybutyryl-S-coenzyme A | HMDB | | Hydroxy-butyryl-CoA | HMDB | | Hydroxy-butyryl-coenzyme A | HMDB | | 3-Hydroxybutyryl-coenzyme A, (R)-isomer | HMDB | | 3-Hydroxybutyryl-coenzyme A, (S)-isomer | HMDB |
|
|---|
| Chemical Formula | C25H42N7O18P3S |
|---|
| Average Molecular Mass | 853.623 g/mol |
|---|
| Monoisotopic Mass | 853.152 g/mol |
|---|
| CAS Registry Number | 2871-66-1 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({hydroxy[3-hydroxy-3-({2-[(2-{[(3R)-3-hydroxybutanoyl]sulfanyl}ethyl)carbamoyl]ethyl}carbamoyl)-2,2-dimethylpropoxy]phosphoryl}oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[({hydroxy[hydroxy(3-hydroxy-3-({2-[(2-{[(3R)-3-hydroxybutanoyl]sulfanyl}ethyl)carbamoyl]ethyl}carbamoyl)-2,2-dimethylpropoxy)phosphoryl]oxyphosphoryl}oxy)methyl]oxolan-3-yl]oxyphosphonic acid |
|---|
| SMILES | C[C@@H](O)CC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C25H42N7O18P3S/c1-13(33)8-16(35)54-7-6-27-15(34)4-5-28-23(38)20(37)25(2,3)10-47-53(44,45)50-52(42,43)46-9-14-19(49-51(39,40)41)18(36)24(48-14)32-12-31-17-21(26)29-11-30-22(17)32/h11-14,18-20,24,33,36-37H,4-10H2,1-3H3,(H,27,34)(H,28,38)(H,42,43)(H,44,45)(H2,26,29,30)(H2,39,40,41)/t13-,14-,18-,19-,20?,24-/m1/s1 |
|---|
| InChI Key | QHHKKMYHDBRONY-JYMPOPDUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as (r)-3-hydroxyacyl coas. These are organic compounds containing a (R)-3-hydroxyl acylated coenzyme A derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | (R)-3-hydroxyacyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0931000130-d924499294c56a627b62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-1912000000-416f64a8b75913d6ee9c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1920000000-137c383401c378170f3d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-053r-9720041570-1667d7f76a22ea47025f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-5910220010-688de5c2366fa2409e73 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-7900100000-b81dd46468dec5a6d312 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001166 |
|---|
| FooDB ID | FDB022460 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | 36912 |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | 456 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 389056 |
|---|
| ChEBI ID | 15452 |
|---|
| PubChem Compound ID | 440045 |
|---|
| Kegg Compound ID | C03561 |
|---|
| YMDB ID | YMDB16133 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|