| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:26:08 UTC |
|---|
| Update Date | 2016-11-09 01:21:14 UTC |
|---|
| Accession Number | CHEM035036 |
|---|
| Identification |
|---|
| Common Name | Phosphohydroxypyruvic acid |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
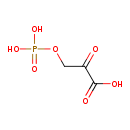
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Phosphohydroxypyruvic acid | ChEBI | | Hydroxypyruvic acid phosphate | ChEBI | | 3-Phosphonooxypyruvic acid | Kegg | | 3-Phosphohydroxypyruvate | Kegg | | 3-Phosphooxypyruvate | Kegg | | Hydroxypyruvate phosphate | Generator | | Hydroxypyruvic acid phosphoric acid | Generator | | 3-Phosphonooxypyruvate | Generator | | 3-Phosphooxypyruvic acid | Generator | | Phosphohydroxypyruvate | Generator | | 2-oxo-3-(Phosphonooxy)-propanoate | HMDB | | 2-oxo-3-(Phosphonooxy)-propanoic acid | HMDB | | 3-Phosphonatooxypyruvate | HMDB | | 2-Oxo-3-(phosphonooxy)propanoic acid | HMDB | | 2-Oxo-3-(phosphonooxy)propionic acid | HMDB | | Phosphohydroxypyruvic acid | HMDB |
|
|---|
| Chemical Formula | C3H5O7P |
|---|
| Average Molecular Mass | 184.041 g/mol |
|---|
| Monoisotopic Mass | 183.977 g/mol |
|---|
| CAS Registry Number | 3913-50-6 |
|---|
| IUPAC Name | 2-oxo-3-(phosphonooxy)propanoic acid |
|---|
| Traditional Name | phosphohydroxypyruvate |
|---|
| SMILES | OC(=O)C(=O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H5O7P/c4-2(3(5)6)1-10-11(7,8)9/h1H2,(H,5,6)(H2,7,8,9) |
|---|
| InChI Key | LFLUCDOSQPJJBE-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Glycerone phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycerone phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Monosaccharide
- Keto acid
- Alpha-keto acid
- Alpha-hydroxy ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9300000000-33ff06735f8214248ea3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00xr-9510000000-f67b2feb11cbb37fdf47 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-4411ba8f6713e6b68f94 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00s9-0900000000-457227604cff5cdecb73 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05g0-6900000000-5ced177e050ba3272f0f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-3900000000-22820af5ec79da81207b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9300000000-0127ef53c9426c592779 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4ba77bd112070310c576 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-1900000000-0d44a3e3fb04adf7390b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000f-9400000000-29a2afefebab0a9dc88b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0016-9000000000-2f054e3a4d1b04f903f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9500000000-d989c4c4b7671c11809a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-7e6d85bfb63dfb4e3ea0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001024 |
|---|
| FooDB ID | FDB022377 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00019657 |
|---|
| BiGG ID | 41453 |
|---|
| BioCyc ID | 3-P-HYDROXYPYRUVATE |
|---|
| METLIN ID | 484 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Phosphohydroxypyruvic acid |
|---|
| Chemspider ID | 103 |
|---|
| ChEBI ID | 30933 |
|---|
| PubChem Compound ID | 105 |
|---|
| Kegg Compound ID | C03232 |
|---|
| YMDB ID | YMDB00393 |
|---|
| ECMDB ID | ECMDB01024 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Zhao G, Winkler ME: A novel alpha-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol. 1996 Jan;178(1):232-9. | | 2. Achouri Y, Rider MH, Schaftingen EV, Robbi M: Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem J. 1997 Apr 15;323 ( Pt 2):365-70. | | 3. Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, Thorleifsson SG, Agren R, Bolling C, Bordel S, Chavali AK, Dobson P, Dunn WB, Endler L, Hala D, Hucka M, Hull D, Jameson D, Jamshidi N, Jonsson JJ, Juty N, Keating S, Nookaew I, Le Novere N, Malys N, Mazein A, Papin JA, Price ND, Selkov E Sr, Sigurdsson MI, Simeonidis E, Sonnenschein N, Smallbone K, Sorokin A, van Beek JH, Weichart D, Goryanin I, Nielsen J, Westerhoff HV, Kell DB, Mendes P, Palsson BO: A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013 May;31(5):419-25. doi: 10.1038/nbt.2488. Epub 2013 Mar 3. |
|
|---|