| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:20:20 UTC |
|---|
| Update Date | 2016-11-09 01:21:13 UTC |
|---|
| Accession Number | CHEM034927 |
|---|
| Identification |
|---|
| Common Name | Coproporphyrin IV |
|---|
| Class | Small Molecule |
|---|
| Description | Coproporhyrin IV is a porphyrin metabolite arising from heme synthesis. Porphyrins are pigments found in both animal and plant life. This is a rare coproporphyrin isomer and it constitues only 2% of all coproporphyrins. This isomer appears to arise spontaneously as opposed to enzymatically.
Coproporphyrin IV is a tetrapyrrole dead-end product from the spontaneous oxidation of the methylene bridges of coproporphynogen, arising from heme synthesis and secreted in feces and urine. Increased levels of coproporphyrins can indicate congenital erythropoietic porphyria or sideroblastic anaemia. Porphyria is a pathological state characterised by abnormalities of porphyrin metabolism and results in the excretion of large quantities of porphyrins in the urine and in extreme sensitivity to light. A large number of factors are capable of increasing porphyrin excretion, owing to different and multiple causes and etiologies: 1) the main site of the chronic hepatic porphyria disease process concentrates on the liver, 2) a functional and morphologic liver injury is almost regularly associated with this chronic porphyria, 3) the toxic form due to occupational and environmental exposure takes mainly a subclinical course.
Hepatic factors includes disturbance in coproporphyrinogen metabolism, which results from inhibition of coproporphyrinogen oxidase as well as from the rapid loss from, and diminished utilization of coproporphyrinogen in the hepatocytes, which may also explain why coproporphyrin, its autoxidation product, predominates physiologically in the urine; decreased biliary excretion of coproporphyrin leading to a compensatory urinary excretion, so that the coproporphyrin ring isomer ratio (1:III) becomes a sensitive index for impaired liver function and intrahepatic cholestasis; and disturbed activity of hepatic uroporphyrinogen decarboxylase. In itself, secondary coproporphyrinuria is not associated with porphyria symptoms of a hepatologic-gastroenterologic, neurologic, or dermatologic order, even though coproporphyrinuria can occur with such symptoms. (PMID: 3327428) [HMDB] |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
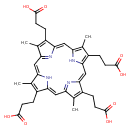
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,8,12,17-Tetramethyl-2,7,13,18-porphinetetrapropionate | HMDB | | 3,8,12,17-Tetramethyl-2,7,13,18-porphinetetrapropionic acid | HMDB | | 3-[10,15,19-Tris(2-carboxyethyl)-5,9,14,20-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1³,⁶.1⁸,¹¹.1¹³,¹⁶]tetracosa-1(21),2,4,6,8(23),9,11,13,15,17,19-undecaen-4-yl]propanoate | Generator, HMDB |
|
|---|
| Chemical Formula | C36H38N4O8 |
|---|
| Average Molecular Mass | 654.709 g/mol |
|---|
| Monoisotopic Mass | 654.269 g/mol |
|---|
| CAS Registry Number | 18372-11-7 |
|---|
| IUPAC Name | 3-[10,15,19-tris(2-carboxyethyl)-5,9,14,20-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1(20),2,4,6(24),7,9,11,13(22),14,16,18-undecaen-4-yl]propanoic acid |
|---|
| Traditional Name | 3-[10,15,19-tris(2-carboxyethyl)-5,9,14,20-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1(20),2,4,6(24),7,9,11,13(22),14,16,18-undecaen-4-yl]propanoic acid |
|---|
| SMILES | CC1=C(CCC(O)=O)/C2=C/C3=N/C(=C\C4=C(CCC(O)=O)C(C)=C(N4)/C=C4\N=C(\C=C\1/N\2)C(C)=C4CCC(O)=O)/C(CCC(O)=O)=C3C |
|---|
| InChI Identifier | InChI=1S/C36H38N4O8/c1-17-21(5-9-33(41)42)29-14-27-19(3)23(7-11-35(45)46)31(39-27)16-32-24(8-12-36(47)48)20(4)28(40-32)15-30-22(6-10-34(43)44)18(2)26(38-30)13-25(17)37-29/h13-16,37,40H,5-12H2,1-4H3,(H,41,42)(H,43,44)(H,45,46)(H,47,48)/b25-13-,26-13-,27-14-,28-15-,29-14-,30-15-,31-16-,32-16- |
|---|
| InChI Key | WEUFSMQSXVJGBC-UJJXFSCMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Tetrapyrroles and derivatives |
|---|
| Sub Class | Porphyrins |
|---|
| Direct Parent | Porphyrins |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052g-2000079000-db9e035127d070b66656 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0000029000-d0154577b253d5531a21 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05p7-0000095000-65e946b9f6e63f6990f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ot-0000090000-374e957ca923df0d583b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0000019000-058ac9e2f7ada938b89a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0k9l-1000049000-123f05a57ae390b6b5a5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9000063000-e208ac274a0e1f580adc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05n0-0000029000-abd7624a794a6460279a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05my-0000096000-5e6abaecc82c2a8c8051 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-0000091000-29869e69aacdfb9e7b08 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udr-0000019000-4aec3c7630d4f4075788 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00ko-0000093000-d5029ff99f55e249d612 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03di-0000090000-7b7f75057f7427120af1 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0000644 |
|---|
| FooDB ID | FDB022160 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 16736703 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | Not Available |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | Not Available |
|---|