| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:15:24 UTC |
|---|
| Update Date | 2016-11-09 01:21:11 UTC |
|---|
| Accession Number | CHEM034827 |
|---|
| Identification |
|---|
| Common Name | 1,3,12-Trihydroxycholan-24-oic acid |
|---|
| Class | Small Molecule |
|---|
| Description | PG(18:0/20:3(8Z,11Z,14Z)) is a phosphatidylglycerol or glycerophospholipid (PG or GP). It is a glycerophospholipid in which a phosphoglycerol moiety occupies a glycerol substitution site. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached at the C-1 and C-2 positions. Fatty acids containing 16, 18 and 20 carbons are the most common. PG(18:0/20:3(8Z,11Z,14Z)), in particular, consists of one chain of stearic acid at the C-1 position and one chain of homo-g-linolenic acid at the C-2 position. The stearic acid moiety is derived from animal fats, coco butter and sesame oil, while the homo-g-linolenic acid moiety is derived from fish oils, liver and kidney. Phosphatidylglycerol is present at a level of 1-2% in most animal tissues, but it can be the second most abundant phospholipid in lung surfactant at up to 11% of the total. It is well established that the concentration of phosphatidylglycerol increases during fetal development. Phosphatidylglycerol may be present in animal tissues merely as a precursor for diphosphatidylglycerol (cardiolipin). Phosphatidylglycerol is formed from phosphatidic acid by a sequence of enzymatic reactions that proceeds via the intermediate, cytidine diphosphate diacylglycerol (CDP-diacylglycerol). Bioynthesis proceeds by condensation of phosphatidic acid and cytidine triphosphate with elimination of pyrophosphate via the action of phosphatidate cytidyltransferase (or CDP-synthase). CDP-diacylglycerol then reacts with glycerol-3-phosphate via phosphatidylglycerophosphate synthase to form 3-sn-phosphatidyl-1'-sn-glycerol 3'-phosphoric acid, with the release of cytidine monophosphate (CMP). Finally, phosphatidylglycerol is formed by the action of specific phosphatases.
While most phospholipids have a saturated fatty acid on C-1 and an unsaturated fatty acid on C-2 of the glycerol backbone, the fatty acid distribution at the C-1 and C-2 positions of glycerol within phospholipids is continually in flux, owing to phospholipid degradation and the continuous phospholipid remodeling that occurs while these molecules are in membranes. PGs have a net charge of -1 at physiological pH and are found in high concentration in mitochondrial membranes and as components of pulmonary surfactant. PG also serves as a precursor for the synthesis of cardiolipin. PG is synthesized from CDP-diacylglycerol and glycerol-3-phosphate. [HMDB] |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
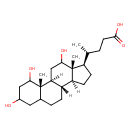
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3,12-Trihydroxy-5b-cholanoate | HMDB | | 1,3,12-Trihydroxy-5b-cholanoic acid | HMDB | | 1,3,12-Trihydroxy-cholan-24-Oate | HMDB | | 1,3,12-Trihydroxy-cholan-24-Oic acid | HMDB | | 1,3,12-Trihydroxycholan-24-Oate | HMDB, Generator | | 1 beta,3 alpha,12 alpha-Trihydroxy-5 beta-cholanoic acid | MeSH, HMDB | | 1,3,12-Trihydroxycholanoic acid | MeSH, HMDB | | (4R)-4-[(1S,2S,10S,11S,14R,15R)-3,5,16-Trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoate | Generator, HMDB |
|
|---|
| Chemical Formula | C24H40O5 |
|---|
| Average Molecular Mass | 408.571 g/mol |
|---|
| Monoisotopic Mass | 408.288 g/mol |
|---|
| CAS Registry Number | 63266-91-1 |
|---|
| IUPAC Name | (4R)-4-[(1S,2S,10S,11S,14R,15R)-3,5,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | (4R)-4-[(1S,2S,10S,11S,14R,15R)-3,5,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoic acid |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)CCC(O)=O)[C@@]1(C)C(O)C[C@@]1([H])[C@@]2([H])CCC2CC(O)CC(O)[C@]12C |
|---|
| InChI Identifier | InChI=1S/C24H40O5/c1-13(4-9-22(28)29)17-7-8-18-16-6-5-14-10-15(25)11-20(26)23(14,2)19(16)12-21(27)24(17,18)3/h13-21,25-27H,4-12H2,1-3H3,(H,28,29)/t13-,14?,15?,16+,17-,18+,19+,20?,21?,23+,24-/m1/s1 |
|---|
| InChI Key | DAKYVYUAVGJDRK-JZBKOBKOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylglycerols. These are glycerophosphoglycerols in which two fatty acids are bonded to the 1-glycerol moiety through ester linkages. As is the case with diacylglycerols, phosphatidylglycerols can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoglycerols |
|---|
| Direct Parent | Phosphatidylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-diacylglycerophosphoglycerol
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- 1,2-diol
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Organic oxide
- Organooxygen compound
- Alcohol
- Organic oxygen compound
- Primary alcohol
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dl-0129000000-93342685f3ac80b739ef | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-001i-1010049000-b3e7856d53a8de4e67bf | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0009000000-0c122eee1b1dbeb1519d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0009000000-06969fbf9e69cbf7a0c1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-3129000000-56243498de002b81b85e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4r-0009800000-4a0ab06b262a99f18ff4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052r-0009200000-9a24af3f58b590ac86cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9008000000-a912af299a7dc32524d7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fu-0009400000-ff2767ac04483681d132 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-059l-3149100000-a3dc8d53445fbc4a7996 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9870000000-0ab9e32eda3ce672b1fe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-ec1d9319a9eaf1f2aba6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-0009800000-986af7877e34b89e542a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gw1-1019100000-fe9fc0214b53ec36a531 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| FooDB ID | FDB027759 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | Not Available |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | Not Available |
|---|