| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 05:06:14 UTC |
|---|
| Update Date | 2016-11-09 01:21:10 UTC |
|---|
| Accession Number | CHEM034661 |
|---|
| Identification |
|---|
| Common Name | 3b,6a-Dihydroxy-alpha-ionol 9-[apiosyl-(1->6)-glucoside] |
|---|
| Class | Small Molecule |
|---|
| Description | 3b,6a-Dihydroxy-alpha-ionol 9-[apiosyl-(1->6)-glucoside] is found in fruits. 3b,6a-Dihydroxy-alpha-ionol 9-[apiosyl-(1->6)-glucoside] is a constituent of Cydonia vulgaris (quince). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
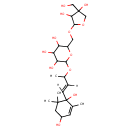
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3b,6a-Dihydroxy-a-ionol 9-[apiosyl-(1->6)-glucoside] | Generator | | 3b,6a-Dihydroxy-α-ionol 9-[apiosyl-(1->6)-glucoside] | Generator |
|
|---|
| Chemical Formula | C24H40O12 |
|---|
| Average Molecular Mass | 520.572 g/mol |
|---|
| Monoisotopic Mass | 520.252 g/mol |
|---|
| CAS Registry Number | 177261-72-2 |
|---|
| IUPAC Name | 2-{[(3E)-4-(1,4-dihydroxy-2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-yl]oxy}-6-({[3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy}methyl)oxane-3,4,5-triol |
|---|
| Traditional Name | 2-{[(3E)-4-(1,4-dihydroxy-2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-yl]oxy}-6-({[3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy}methyl)oxane-3,4,5-triol |
|---|
| SMILES | Not Available |
|---|
| InChI Identifier | InChI=1S/C24H40O12/c1-12-7-14(26)8-22(3,4)24(12,32)6-5-13(2)35-20-18(29)17(28)16(27)15(36-20)9-33-21-19(30)23(31,10-25)11-34-21/h5-7,13-21,25-32H,8-11H2,1-4H3/b6-5+ |
|---|
| InChI Key | FCEJNLVNFSUDTK-AATRIKPKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Sesquiterpenoid
- Megastigmane sesquiterpenoid
- Cyclofarsesane sesquiterpenoid
- Ionone derivative
- Alkyl glycoside
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Oxane
- Tetrahydrofuran
- Tertiary alcohol
- Secondary alcohol
- Polyol
- Oxacycle
- Organoheterocyclic compound
- Acetal
- Primary alcohol
- Organooxygen compound
- Organic oxygen compound
- Alcohol
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zg0-9361670000-0d1e0f0b01981f7514ce | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-000t-2441119000-c552136078d0f3dfde43 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0pk9-0292360000-f20f9d525e9dd3213654 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-1391000000-a8bf55613b98078ffc3e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-6790000000-f8f556d84f1c0424047f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0699-0792560000-b8cf7ebb8a6a564498f9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-0591210000-cbbc4f90b0a190383c93 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-3790000000-023a27093310190661bb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0110190000-b6b63d3ea3ae7ebf2822 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xr-7935210000-066a852e35a6cd30d100 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pdj-7951000000-1bc12cd2a6179b31acc8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0962430000-9eeaa904437426cabc62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-1797620000-068fbab83c6b797c1cae | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pvi-9453000000-769a8ef65f00a7bb1f4e | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041577 |
|---|
| FooDB ID | FDB021568 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 167960 |
|---|
| PubChem Compound ID | 131801688 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|