| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:56:07 UTC |
|---|
| Update Date | 2016-11-09 01:21:08 UTC |
|---|
| Accession Number | CHEM034462 |
|---|
| Identification |
|---|
| Common Name | Gibberellin A100 |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of sunflower (Helianthus annuus) seeds. Gibberellin A100 is found in sunflower and fats and oils. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
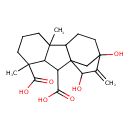
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GA100 | HMDB | | 12,14-Dihydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylate | Generator |
|
|---|
| Chemical Formula | C20H28O6 |
|---|
| Average Molecular Mass | 364.433 g/mol |
|---|
| Monoisotopic Mass | 364.189 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 12,14-dihydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylic acid |
|---|
| Traditional Name | 12,14-dihydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylic acid |
|---|
| SMILES | CC12CCCC(C)(C1C(C(O)=O)C13CC(O)(CCC21)C(=C)C3O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H28O6/c1-10-14(21)20-9-19(10,26)8-5-11(20)17(2)6-4-7-18(3,16(24)25)13(17)12(20)15(22)23/h11-14,21,26H,1,4-9H2,2-3H3,(H,22,23)(H,24,25) |
|---|
| InChI Key | GWJAUARFGPKLMT-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as c20-gibberellin 6-carboxylic acids. These are c20-gibberellins with a carboxyl group at the 6-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | C20-gibberellin 6-carboxylic acids |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000b-0239000000-df54a6407871e50156e8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-000i-3020089000-03382f103f4b534039ec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0009000000-d8b1d9d68f0f3dea6359 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ufs-0019000000-162b1623c01369667758 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-3379000000-2b479e58382ee4c77930 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03xr-0009000000-db4c4edcf76fcdffe515 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uxr-0019000000-cb7472fdc1e5d230970a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zi0-3069000000-fdf634f376c9f46a98fa | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041348 |
|---|
| FooDB ID | FDB021271 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000324 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 14160543 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|