| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:51:35 UTC |
|---|
| Update Date | 2016-11-09 01:21:06 UTC |
|---|
| Accession Number | CHEM034359 |
|---|
| Identification |
|---|
| Common Name | 3-Oxo-8b-hydroxy-7(11)-eremophilen-12,8-olide |
|---|
| Class | Small Molecule |
|---|
| Description | (8betaOH,10beta)-8-Hydroxy-3-oxo-7(11)-eremophilen-12,8-olide is found in green vegetables. (8betaOH,10beta)-8-Hydroxy-3-oxo-7(11)-eremophilen-12,8-olide is a constituent of Petasites japonicus (sweet coltsfoot). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
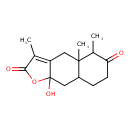
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (8BetaOH,10b)-8-hydroxy-3-oxo-7(11)-eremophilen-12,8-olide | Generator | | (8BetaOH,10β)-8-hydroxy-3-oxo-7(11)-eremophilen-12,8-olide | Generator |
|
|---|
| Chemical Formula | C15H20O4 |
|---|
| Average Molecular Mass | 264.317 g/mol |
|---|
| Monoisotopic Mass | 264.136 g/mol |
|---|
| CAS Registry Number | 171422-92-7 |
|---|
| IUPAC Name | 9a-hydroxy-3,4a,5-trimethyl-2H,4H,4aH,5H,6H,7H,8H,8aH,9H,9aH-naphtho[2,3-b]furan-2,6-dione |
|---|
| Traditional Name | 9a-hydroxy-3,4a,5-trimethyl-4H,5H,7H,8H,8aH,9H-naphtho[2,3-b]furan-2,6-dione |
|---|
| SMILES | CC1C(=O)CCC2CC3(O)OC(=O)C(C)=C3CC12C |
|---|
| InChI Identifier | InChI=1S/C15H20O4/c1-8-11-7-14(3)9(2)12(16)5-4-10(14)6-15(11,18)19-13(8)17/h9-10,18H,4-7H2,1-3H3 |
|---|
| InChI Key | AQAVKEZEFHFROC-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as terpene lactones. These are prenol lipids containing a lactone ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Terpene lactones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Terpene lactone
- Eremophilanolide or secoeremophilanolide
- Sesquiterpenoid
- Naphthofuran
- 2-furanone
- Cyclic alcohol
- Dihydrofuran
- Enoate ester
- Alpha,beta-unsaturated carboxylic ester
- Lactone
- Ketone
- Hemiacetal
- Carboxylic acid ester
- Cyclic ketone
- Oxacycle
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01w3-0950000000-18289cd94d16aedeaffb | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00g0-7793000000-c24cc26b4d113119b8d6 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0390000000-d0bc0769a430e579c111 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kv-0960000000-0f5f8eb2aa4569392410 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gc0-9400000000-319a711b1ef0e9a07928 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0090000000-d2c7110fd048769b8f9e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-07vi-0090000000-a9af63e8ac1435b7b0c1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05mo-6690000000-f2e4407556fb71764510 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0090000000-16d43704d08f3756e03a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0090000000-a20608b0aca0e1e4b9a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-07p1-1390000000-7d71ac7c8c3459c7f4a2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0090000000-27f9036d868b151212c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01b9-0790000000-0d22e1c5a38532010230 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00or-5910000000-c5fee28880a0630db3c9 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041227 |
|---|
| FooDB ID | FDB021129 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35015132 |
|---|
| ChEBI ID | 174485 |
|---|
| PubChem Compound ID | 85231437 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|