| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:47:49 UTC |
|---|
| Update Date | 2016-11-09 01:21:05 UTC |
|---|
| Accession Number | CHEM034273 |
|---|
| Identification |
|---|
| Common Name | Sonchifolin |
|---|
| Class | Small Molecule |
|---|
| Description | Sonchifolin is found in green vegetables. Sonchifolin is a constituent of Smallanthus sonchifolius (yacon). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
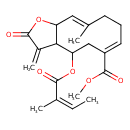
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Methyl 10-methyl-4-{[(2Z)-2-methylbut-2-enoyl]oxy}-3-methylidene-2-oxo-2H,3H,3ah,4H,5H,8H,9H,11ah-cyclodeca[b]furan-6-carboxylic acid | HMDB |
|
|---|
| Chemical Formula | C21H26O6 |
|---|
| Average Molecular Mass | 374.428 g/mol |
|---|
| Monoisotopic Mass | 374.173 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | methyl 10-methyl-4-{[(2Z)-2-methylbut-2-enoyl]oxy}-3-methylidene-2-oxo-2H,3H,3aH,4H,5H,8H,9H,11aH-cyclodeca[b]furan-6-carboxylate |
|---|
| Traditional Name | methyl 10-methyl-4-{[(2Z)-2-methylbut-2-enoyl]oxy}-3-methylidene-2-oxo-3aH,4H,5H,8H,9H,11aH-cyclodeca[b]furan-6-carboxylate |
|---|
| SMILES | COC(=O)C1=C/CC\C(C)=C\C2OC(=O)C(=C)C2C(C\1)OC(=O)C(\C)=C/C |
|---|
| InChI Identifier | InChI=1S/C21H26O6/c1-6-13(3)19(22)26-17-11-15(21(24)25-5)9-7-8-12(2)10-16-18(17)14(4)20(23)27-16/h6,9-10,16-18H,4,7-8,11H2,1-3,5H3/b12-10+,13-6-,15-9+ |
|---|
| InChI Key | DBSZDVZLFLCIOQ-VPXSVSNXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as germacranolides and derivatives. These are sesquiterpene lactones with a structure based on the germacranolide skeleton, characterized by a gamma lactone fused to a 1,7-dimethylcyclodec-1-ene moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Germacranolides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Germacranolide
- Germacrane sesquiterpenoid
- Sesquiterpenoid
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Gamma butyrolactone
- Fatty acyl
- Tetrahydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Methyl ester
- Enoate ester
- Lactone
- Carboxylic acid ester
- Organoheterocyclic compound
- Oxacycle
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053r-9012000000-1086956d858c12b3217a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-2029000000-6951c1877f1630292970 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-057i-9064000000-292e922cdb404c64ce75 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05o0-9030000000-69fa9c6974dbc6910c4b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0019000000-ef1a928d89896f708bce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05i4-4039000000-fefcf7ce3b07625f20c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053b-5091000000-8036a13a030e3c53f076 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9054000000-909b651c8de505ec7061 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052b-9081000000-6708d0002669c7d3a979 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-8090000000-55569ccb60ecf1b1a9fa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0091000000-02a14b321b75f7f682d2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0091000000-536d62f6a539e6e709d6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ldi-7190000000-3fe11a28060f795315d4 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041139 |
|---|
| FooDB ID | FDB021024 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35015114 |
|---|
| ChEBI ID | 175820 |
|---|
| PubChem Compound ID | 131753040 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|