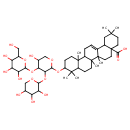| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:35:11 UTC |
|---|
| Update Date | 2016-11-09 01:21:02 UTC |
|---|
| Accession Number | CHEM033994 |
|---|
| Identification |
|---|
| Common Name | Elatoside E |
|---|
| Class | Small Molecule |
|---|
| Description | Elatoside E is found in green vegetables. Elatoside E is a constituent of Aralia elata (Japanese angelica tree). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Tarasaponin III | HMDB | | 10-[(5-Hydroxy-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3-[(3,4,5-trihydroxyoxan-2-yl)oxy]oxan-2-yl)oxy]-2,2,6a,6b,9,9,12a-heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylate | Generator | | Elatoside e | MeSH |
|
|---|
| Chemical Formula | C46H74O16 |
|---|
| Average Molecular Mass | 883.070 g/mol |
|---|
| Monoisotopic Mass | 882.498 g/mol |
|---|
| CAS Registry Number | 156980-30-2 |
|---|
| IUPAC Name | 10-[(5-hydroxy-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3-[(3,4,5-trihydroxyoxan-2-yl)oxy]oxan-2-yl)oxy]-2,2,6a,6b,9,9,12a-heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylic acid |
|---|
| Traditional Name | 10-[(5-hydroxy-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3-[(3,4,5-trihydroxyoxan-2-yl)oxy]oxan-2-yl)oxy]-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,7,8,8a,10,11,12,12b,13,14b-tetradecahydropicene-4a-carboxylic acid |
|---|
| SMILES | CC1(C)CCC2(CCC3(C)C(=CCC4C5(C)CCC(OC6OCC(O)C(OC7OC(CO)C(O)C(O)C7O)C6OC6OCC(O)C(O)C6O)C(C)(C)C5CCC34C)C2C1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C46H74O16/c1-41(2)14-16-46(40(55)56)17-15-44(6)22(23(46)18-41)8-9-28-43(5)12-11-29(42(3,4)27(43)10-13-45(28,44)7)60-39-36(62-37-33(53)30(50)24(48)20-57-37)35(25(49)21-58-39)61-38-34(54)32(52)31(51)26(19-47)59-38/h8,23-39,47-54H,9-21H2,1-7H3,(H,55,56) |
|---|
| InChI Key | QISCHUABGXFSHX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Oxane
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Oxacycle
- Polyol
- Organoheterocyclic compound
- Organic oxide
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Primary alcohol
- Organic oxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-107r-0100710940-de0512d113253215373a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052r-0100940500-d43ad52966901c951db0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052r-2212970310-70322342bc90d53f568a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001j-1701602890-1c25558960698996c46d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06rt-0800921720-842ec2c13018722f6ccc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4m-7700920000-3d6aad2be2509f653c86 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0101200190-ad47b0bfdf4e36bf4688 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0016-0649331360-ac1a63c366b01a0cc189 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0081-2920011320-cfeb08af4dbe1d316e15 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0100000290-7910751c32bd3638791f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-9501200570-9ea557ae37ca2450ce41 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dl-7100090300-efb2b806b5e396a98dfa | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040852 |
|---|
| FooDB ID | FDB020677 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00057940 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 78174063 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|