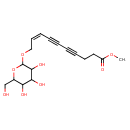| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:32:37 UTC |
|---|
| Update Date | 2016-11-09 01:21:02 UTC |
|---|
| Accession Number | CHEM033939 |
|---|
| Identification |
|---|
| Common Name | Methyl helianthenoate F glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | Methyl helianthenoate F glucoside is found in root vegetables. Methyl helianthenoate F glucoside is a constituent of Helianthus tuberosus (Jerusalem artichoke). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Methyl helianthenoic acid F glucoside | Generator | | Methyl (8Z)-10-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}dec-8-en-4,6-diynoic acid | Generator |
|
|---|
| Chemical Formula | C17H22O8 |
|---|
| Average Molecular Mass | 354.352 g/mol |
|---|
| Monoisotopic Mass | 354.131 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | methyl (8Z)-10-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}dec-8-en-4,6-diynoate |
|---|
| Traditional Name | methyl (8Z)-10-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}dec-8-en-4,6-diynoate |
|---|
| SMILES | COC(=O)CCC#CC#C\C=C/COC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C17H22O8/c1-23-13(19)9-7-5-3-2-4-6-8-10-24-17-16(22)15(21)14(20)12(11-18)25-17/h6,8,12,14-18,20-22H,7,9-11H2,1H3/b8-6- |
|---|
| InChI Key | KMCLOJYUKASTFN-VURMDHGXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Alkyl glycoside
- Hexose monosaccharide
- Glycosyl compound
- O-glycosyl compound
- Fatty acid ester
- Sugar acid
- Fatty acid methyl ester
- Oxane
- Monosaccharide
- Methyl ester
- Secondary alcohol
- Carboxylic acid ester
- Polyol
- Carboxylic acid derivative
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Oxacycle
- Acetal
- Primary alcohol
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Organic oxygen compound
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0079-6955000000-ea99596cc93e5113efe5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-004i-2311239000-a2bb99c5364ec7e6f144 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0abi-0309000000-1ef915e633db3fed1f5e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01tc-1901000000-84f6fb9106b59c927743 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00mo-9720000000-943b61f266fbab574567 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-1519000000-2084f289120d05b1c2d7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03mu-4913000000-9cbd4540c4230d35b76b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9510000000-a22d108293e63f09ad35 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-1093-0902000000-a9eb8d60b49032a72709 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gbi-3900000000-c9a177d71559fb8543cb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uy0-2900000000-4441b33a5ee3d0606e11 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0uk9-0219000000-722b9321d6f05ddf6d31 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bt9-3923000000-16e68eab128b8cd1523e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053r-3900000000-e5121341c99446a9658f | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040797 |
|---|
| FooDB ID | FDB020613 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 74886487 |
|---|
| ChEBI ID | 168120 |
|---|
| PubChem Compound ID | 131752941 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|