| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:30:57 UTC |
|---|
| Update Date | 2016-11-09 01:21:01 UTC |
|---|
| Accession Number | CHEM033898 |
|---|
| Identification |
|---|
| Common Name | 3b-Hydroxy-6b,8a-dimethoxy-7(11)-eremophilen-12,8-olide |
|---|
| Class | Small Molecule |
|---|
| Description | 3-Hydroxy-6,8-dimethoxy-7(11)-eremophilen-12,8-olide is found in green vegetables. 3-Hydroxy-6,8-dimethoxy-7(11)-eremophilen-12,8-olide is a constituent of Petasites japonicus (sweet coltsfoot). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
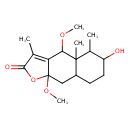
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C17H26O5 |
|---|
| Average Molecular Mass | 310.385 g/mol |
|---|
| Monoisotopic Mass | 310.178 g/mol |
|---|
| CAS Registry Number | 162613-65-2 |
|---|
| IUPAC Name | 6-hydroxy-4,9a-dimethoxy-3,4a,5-trimethyl-2H,4H,4aH,5H,6H,7H,8H,8aH,9H,9aH-naphtho[2,3-b]furan-2-one |
|---|
| Traditional Name | 6-hydroxy-4,9a-dimethoxy-3,4a,5-trimethyl-4H,5H,6H,7H,8H,8aH,9H-naphtho[2,3-b]furan-2-one |
|---|
| SMILES | COC1C2=C(C)C(=O)OC2(CC2CCC(O)C(C)C12C)OC |
|---|
| InChI Identifier | InChI=1S/C17H26O5/c1-9-13-14(20-4)16(3)10(2)12(18)7-6-11(16)8-17(13,21-5)22-15(9)19/h10-12,14,18H,6-8H2,1-5H3 |
|---|
| InChI Key | JCGMJARSAZGXHM-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as terpene lactones. These are prenol lipids containing a lactone ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Terpene lactones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Terpene lactone
- Eremophilanolide or secoeremophilanolide
- Sesquiterpenoid
- Naphthofuran
- Ketal
- 2-furanone
- Cyclic alcohol
- Dihydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Carboxylic acid ester
- Lactone
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Ether
- Oxacycle
- Dialkyl ether
- Acetal
- Carboxylic acid derivative
- Organoheterocyclic compound
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Organooxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-017r-0290000000-2b4031c5a33419f48408 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fr-2159000000-30c6977722f182f456ae | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-0093000000-4ea9d10cbb149ffb1e53 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ikc-2091000000-2394f3ae4fb7aeeb36fc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zi0-3930000000-4b1fd4b6ccaa09102809 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0098000000-194bdfb77213b2a968f3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0btc-0092000000-4f04f58de5082faea610 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-059i-1490000000-a97101c3ff37bddbc212 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-0179000000-30dba68326d91125da38 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0191000000-28513b1d889e45ed8ff9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a5j-2910000000-58cd930c1e2d96a7a703 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0019000000-5e4c9d7c7930f298d336 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0019000000-fb27b2988cf70e4ec1c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-002r-1970000000-625584ee3884136720f8 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040756 |
|---|
| FooDB ID | FDB020570 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35015014 |
|---|
| ChEBI ID | 143054 |
|---|
| PubChem Compound ID | 85094081 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|