| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:26:37 UTC |
|---|
| Update Date | 2016-11-09 01:21:00 UTC |
|---|
| Accession Number | CHEM033803 |
|---|
| Identification |
|---|
| Common Name | 3-trans-Caffeoyltormentic acid |
|---|
| Class | Small Molecule |
|---|
| Description | 3-trans-Caffeoyltormentic acid is found in fruits. 3-trans-Caffeoyltormentic acid is a constituent of Eriobotrya japonica (loquat). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
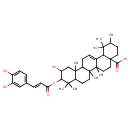
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-trans-Caffeoyltormentate | Generator | | 10-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}-1,11-dihydroxy-1,2,6a,6b,9,9,12a-heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylate | HMDB |
|
|---|
| Chemical Formula | C39H54O8 |
|---|
| Average Molecular Mass | 650.841 g/mol |
|---|
| Monoisotopic Mass | 650.382 g/mol |
|---|
| CAS Registry Number | 144604-16-0 |
|---|
| IUPAC Name | 10-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}-1,11-dihydroxy-1,2,6a,6b,9,9,12a-heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylic acid |
|---|
| Traditional Name | 10-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}-1,11-dihydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,6,7,8,8a,10,11,12,12b,13,14b-tetradecahydropicene-4a-carboxylic acid |
|---|
| SMILES | CC1CCC2(CCC3(C)C(=CCC4C5(C)CC(O)C(OC(=O)\C=C\C6=CC=C(O)C(O)=C6)C(C)(C)C5CCC34C)C2C1(C)O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C39H54O8/c1-22-14-17-39(33(44)45)19-18-36(5)24(31(39)38(22,7)46)10-12-29-35(4)21-27(42)32(34(2,3)28(35)15-16-37(29,36)6)47-30(43)13-9-23-8-11-25(40)26(41)20-23/h8-11,13,20,22,27-29,31-32,40-42,46H,12,14-19,21H2,1-7H3,(H,44,45)/b13-9+ |
|---|
| InChI Key | MFPIVTNHMCQSEM-UKTHLTGXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Cinnamic acid or derivatives
- Coumaric acid or derivatives
- Hydroxycinnamic acid or derivatives
- Cinnamic acid ester
- Catechol
- Styrene
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Fatty acid ester
- Phenol
- Benzenoid
- Fatty acyl
- Monocyclic benzene moiety
- Dicarboxylic acid or derivatives
- Tertiary alcohol
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Cyclic alcohol
- Secondary alcohol
- Carboxylic acid ester
- Carboxylic acid derivative
- Carboxylic acid
- Hydrocarbon derivative
- Alcohol
- Organic oxygen compound
- Organic oxide
- Organooxygen compound
- Carbonyl group
- Aromatic homopolycyclic compound
|
|---|
| Molecular Framework | Aromatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00kr-2203907000-4e4cf95eb47df0db9c1a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_9) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ff0-0300419000-8ee5dddb52b780d11160 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0900836000-793988fa2ca6b51495b4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00g3-1401911000-0d06d690b110050167c9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0200219000-11383b227e5a15c61f9a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kr-0700936000-edeb5978adee2bc08264 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02g6-1600900000-a7e9388cbea1b5f90d6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0100209000-3cb758331b0afbf6384e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p2-0900201000-d5cb35f65406c1b0570e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a5a-9724322000-ff81d7406d9a6b47099e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000209000-1d200903fb00ec4719fd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000j-0700907000-ca0477eb075db3a6e795 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-0900000000-a716d281c1b0bd76d004 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040650 |
|---|
| FooDB ID | FDB020443 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00055127 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014991 |
|---|
| ChEBI ID | 168484 |
|---|
| PubChem Compound ID | 131752884 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|