| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:25:57 UTC |
|---|
| Update Date | 2016-11-09 01:21:00 UTC |
|---|
| Accession Number | CHEM033787 |
|---|
| Identification |
|---|
| Common Name | Piperochromanoic acid |
|---|
| Class | Small Molecule |
|---|
| Description | Piperochromanoic acid is found in herbs and spices. Piperochromanoic acid is a constituent of the leaves of Piper auritum (Veracruz pepper). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
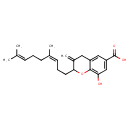
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-(4,8-Dimethyl-3,7-nonadienyl)-3,4-dihydro-8-hydroxy-3-methylene-2H-1-benzopyran-6-carboxylic acid, 9ci | HMDB | | 2-(4,8-Dimethylnona-3,7-dien-1-yl)-8-hydroxy-3-methylidene-3,4-dihydro-2H-1-benzopyran-6-carboxylate | Generator |
|
|---|
| Chemical Formula | C22H28O4 |
|---|
| Average Molecular Mass | 356.455 g/mol |
|---|
| Monoisotopic Mass | 356.199 g/mol |
|---|
| CAS Registry Number | 110979-03-8 |
|---|
| IUPAC Name | 2-[(3Z)-4,8-dimethylnona-3,7-dien-1-yl]-8-hydroxy-3-methylidene-3,4-dihydro-2H-1-benzopyran-6-carboxylic acid |
|---|
| Traditional Name | 2-[(3Z)-4,8-dimethylnona-3,7-dien-1-yl]-8-hydroxy-3-methylidene-2,4-dihydro-1-benzopyran-6-carboxylic acid |
|---|
| SMILES | CC(C)=CCC\C(C)=C/CCC1OC2=C(O)C=C(C=C2CC1=C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C22H28O4/c1-14(2)7-5-8-15(3)9-6-10-20-16(4)11-17-12-18(22(24)25)13-19(23)21(17)26-20/h7,9,12-13,20,23H,4-6,8,10-11H2,1-3H3,(H,24,25)/b15-9- |
|---|
| InChI Key | XBOKPWYVDNVFMX-DHDCSXOGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as bicyclic monoterpenoids. These are monoterpenoids containing exactly 2 rings, which are fused to each other. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Bicyclic monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aromatic monoterpenoid
- Chromane
- Benzopyran
- Bicyclic monoterpenoid
- 1-benzopyran
- Hydroxybenzoic acid
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Alkyl aryl ether
- Benzenoid
- Oxacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Carboxylic acid
- Ether
- Monocarboxylic acid or derivatives
- Organic oxide
- Hydrocarbon derivative
- Organic oxygen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0aor-5695000000-675cd1073a6f17671164 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-014r-6920700000-68baededabf430a45104 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0cdr-0319000000-0b884a131889c6bf1838 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-1911000000-dd13f144965f71f15c72 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-8930000000-652f6e899c7c67b93aca | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0009000000-f0c10b219ae7dbed409d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fr-0309000000-61056d6fc3d4c7d3d566 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ue9-1920000000-a576ae7aedeaaacce726 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-3079000000-8570c2d7184360aa2185 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uyl-7494000000-df29f3a47f62646dfaf7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-5920000000-5c6c0e2b3cff3db659b9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0009000000-a3dd451f5dc822798759 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0319000000-8f76881f8e9b57c0f47d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00mo-0690000000-d8fcba27a4bc3785a3ae | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040634 |
|---|
| FooDB ID | FDB020425 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00056455 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014986 |
|---|
| ChEBI ID | 175587 |
|---|
| PubChem Compound ID | 13845940 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|