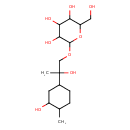| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 04:03:10 UTC |
|---|
| Update Date | 2016-11-09 01:20:53 UTC |
|---|
| Accession Number | CHEM033276 |
|---|
| Identification |
|---|
| Common Name | (1S,2R,4R,8S)-p-Menthane-2,8,9-triol 9-glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | (1S,2R,4R,8S)-p-Menthane-2,8,9-triol 9-glucoside is found in fats and oils. (1S,2R,4R,8S)-p-Menthane-2,8,9-triol 9-glucoside is a constituent of fruit of Carum carvi (caraway). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C16H30O8 |
|---|
| Average Molecular Mass | 350.405 g/mol |
|---|
| Monoisotopic Mass | 350.194 g/mol |
|---|
| CAS Registry Number | 378254-08-1 |
|---|
| IUPAC Name | 2-[2-hydroxy-2-(3-hydroxy-4-methylcyclohexyl)propoxy]-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| Traditional Name | 2-[2-hydroxy-2-(3-hydroxy-4-methylcyclohexyl)propoxy]-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| SMILES | CC1CCC(CC1O)C(C)(O)COC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C16H30O8/c1-8-3-4-9(5-10(8)18)16(2,22)7-23-15-14(21)13(20)12(19)11(6-17)24-15/h8-15,17-22H,3-7H2,1-2H3 |
|---|
| InChI Key | KRTKKVIKRTZKGY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as terpene glycosides. These are prenol lipids containing a carbohydrate moiety glycosidically bound to a terpene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene glycosides |
|---|
| Direct Parent | Terpene glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Terpene glycoside
- Hexose monosaccharide
- Glycosyl compound
- O-glycosyl compound
- P-menthane monoterpenoid
- Monocyclic monoterpenoid
- Monoterpenoid
- Cyclohexanol
- Oxane
- Monosaccharide
- Tertiary alcohol
- Cyclic alcohol
- Secondary alcohol
- Polyol
- Acetal
- Oxacycle
- Organoheterocyclic compound
- Primary alcohol
- Organooxygen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0540-9535000000-dadab6f602201bcf2362 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-4424039000-6b425327fd78c2b19d5d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-0409000000-1d43611a3e0ed4d5d50c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00xr-2902000000-4bebf59c3b97808fbbab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0mbc-6900000000-cf2cf0df296144f6fb1d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1619000000-a20a8d12073faf1683b4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dj-4903000000-1fd5d5cc1143e0631f60 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9800000000-8c6a82dc1289f44090a8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0009000000-4673bb7876c936b20862 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-3539000000-35975fc272de033bd34e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052r-9710000000-f90828491cd68efc69ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fz9-0409000000-03d3e28246b333fd039d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0g4i-2902000000-3ea0e4979146a8e09665 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6y-9310000000-f2de06295d1e798f7731 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039978 |
|---|
| FooDB ID | FDB019653 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 167998 |
|---|
| PubChem Compound ID | 73809749 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|