| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:52:06 UTC |
|---|
| Update Date | 2016-11-09 01:20:50 UTC |
|---|
| Accession Number | CHEM033016 |
|---|
| Identification |
|---|
| Common Name | 25,27-Dihydro-4,7-didehydro-7-deoxyphysalin A |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Physalis alkekengi (winter cherry). 25,27-Dihydro-4,7-didehydro-7-deoxyphysalin A is found in fruits. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
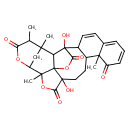
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Etrenol | HMDB | | Hicantona | HMDB | | Hycanthon | HMDB | | Hycanthone | HMDB | | Hycanthone mesylate | HMDB | | Hycanthonum | HMDB | | Lucanthone metabolite | HMDB |
|
|---|
| Chemical Formula | C28H30O9 |
|---|
| Average Molecular Mass | 510.532 g/mol |
|---|
| Monoisotopic Mass | 510.189 g/mol |
|---|
| CAS Registry Number | 134434-71-2 |
|---|
| IUPAC Name | 6,19-dihydroxy-1,15,22,26-tetramethyl-4,21,24-trioxaheptacyclo[21.3.1.0²,⁶.0³,¹⁹.0³,²².0⁷,¹⁶.0¹⁰,¹⁵]heptacosa-8,10,12-triene-5,14,20,25-tetrone |
|---|
| Traditional Name | 6,19-dihydroxy-1,15,22,26-tetramethyl-4,21,24-trioxaheptacyclo[21.3.1.0²,⁶.0³,¹⁹.0³,²².0⁷,¹⁶.0¹⁰,¹⁵]heptacosa-8,10,12-triene-5,14,20,25-tetrone |
|---|
| SMILES | CC1C(=O)OC2CC1(C)C1C3(O)C4C=CC5=CC=CC(=O)C5(C)C4CCC4(O)C(=O)OC2(C)C14OC3=O |
|---|
| InChI Identifier | InChI=1S/C28H30O9/c1-13-19(30)35-18-12-23(13,2)20-27(34)16-9-8-14-6-5-7-17(29)24(14,3)15(16)10-11-26(33)21(31)36-25(18,4)28(20,26)37-22(27)32/h5-9,13,15-16,18,20,33-34H,10-12H2,1-4H3 |
|---|
| InChI Key | NSFHPMKDYVTYOY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as physalins and derivatives. These are steroidal constituents of Physalis plants which possess an unusual 13,14-seco-16,24-cyclo-steroidal ring skeleton (where the bond that is normally present between the 13 and 14 positions in other steroids is broken while a new bond between positions 16 and 24 is formed). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Physalins and derivatives |
|---|
| Direct Parent | Physalins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Physalin skeleton
- Tricarboxylic acid or derivatives
- Delta valerolactone
- Delta_valerolactone
- Oxane
- Gamma butyrolactone
- Tetrahydrofuran
- Tertiary alcohol
- Carboxylic acid ester
- Lactone
- Ketone
- Organoheterocyclic compound
- Oxacycle
- Carboxylic acid derivative
- Hydrocarbon derivative
- Alcohol
- Organooxygen compound
- Organic oxygen compound
- Carbonyl group
- Organic oxide
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01bi-1910800000-ad6602e2aafaa628f9df | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01p9-2700009000-7e9b28b8aa1c1bec62d9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000970000-d4377fc68bdf2d9944cb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ox-0000910000-ce7510f47f063a0e3818 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000j-1002900000-7f210a102c0b8ec77817 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000790000-c7ce07f9a2bb619e7ba1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0aou-0000940000-778a8b3d6268d2693153 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-1801900000-2fafaa2f69b54ce1a34d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000190000-d2b23ee53da864be7d38 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0000390000-c9e9063d703c15dc7a62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0200980000-6396cae14c9dfb368b2e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000090000-1aa2dce78661b19c596b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-0200930000-4fdf8c53d9f12b1ab6b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-1901200000-6d34565309509c23ee2b | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039696 |
|---|
| FooDB ID | FDB019325 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 73825229 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|