| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:49:06 UTC |
|---|
| Update Date | 2016-11-09 01:20:49 UTC |
|---|
| Accession Number | CHEM032943 |
|---|
| Identification |
|---|
| Common Name | Gibberellin A90 |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Triticum aestivum (wheat). Gibberellin A90 is found in wheat, cereals and cereal products, and common wheat. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
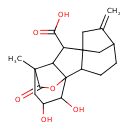
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GA90 | HMDB | | 13,14-Dihydroxy-11-methyl-6-methylidene-16-oxo-15-oxapentacyclo[9.3.2.1⁵,⁸.0¹,¹⁰.0²,⁸]heptadecane-9-carboxylate | Generator |
|
|---|
| Chemical Formula | C19H24O6 |
|---|
| Average Molecular Mass | 348.390 g/mol |
|---|
| Monoisotopic Mass | 348.157 g/mol |
|---|
| CAS Registry Number | 152110-34-4 |
|---|
| IUPAC Name | 13,14-dihydroxy-11-methyl-6-methylidene-16-oxo-15-oxapentacyclo[9.3.2.1⁵,⁸.0¹,¹⁰.0²,⁸]heptadecane-9-carboxylic acid |
|---|
| Traditional Name | 13,14-dihydroxy-11-methyl-6-methylidene-16-oxo-15-oxapentacyclo[9.3.2.1⁵,⁸.0¹,¹⁰.0²,⁸]heptadecane-9-carboxylic acid |
|---|
| SMILES | CC12CC(O)C(O)C3(OC1=O)C1CCC4CC1(CC4=C)C(C23)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C19H24O6/c1-8-5-18-6-9(8)3-4-11(18)19-13(12(18)15(22)23)17(2,16(24)25-19)7-10(20)14(19)21/h9-14,20-21H,1,3-7H2,2H3,(H,22,23) |
|---|
| InChI Key | ZRNQDBNUGCCKBO-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as c19-gibberellin 6-carboxylic acids. These are c19-gibberellins with a carboxyl group at the 6-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | C19-gibberellin 6-carboxylic acids |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fsl-7609000000-0f2d179a40e43f27c143 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-05dj-6410590000-73ead1f9d7c200af6906 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000t-0009000000-027ff5062af796647bf9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fss-0549000000-b46bee983ecfa9fb358e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014r-4961000000-aca5bb744c00c2fd5530 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0029000000-9f800acca7c765de09f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udj-0049000000-56d2585eae2ae3390f6e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-1492000000-8e941f8cb085d2a52048 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039622 |
|---|
| FooDB ID | FDB019250 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000090 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 85370845 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|