| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:41:22 UTC |
|---|
| Update Date | 2016-11-09 01:20:47 UTC |
|---|
| Accession Number | CHEM032772 |
|---|
| Identification |
|---|
| Common Name | (2alpha,3alpha,5alpha,22R,23R)-2,3,22,23-Tetrahydroxy-25-methylergost-24(28)en-6-one |
|---|
| Class | Small Molecule |
|---|
| Description | (2beta,3beta,5alpha,22R,23R)-2,3,22,23-Tetrahydroxy-25-methylergost-24(28)en-6-one is found in common bean. (2beta,3beta,5alpha,22R,23R)-2,3,22,23-Tetrahydroxy-25-methylergost-24(28)en-6-one is a constituent of the immature seeds of Phaseolus vulgaris (kidney bean). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
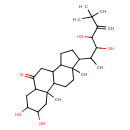
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2a,3a,5a,22R,23R)-2,3,22,23-Tetrahydroxy-25-methylergost-24(28)en-6-one | Generator | | (2Α,3α,5α,22R,23R)-2,3,22,23-tetrahydroxy-25-methylergost-24(28)en-6-one | Generator | | 25-Methyldiolichosterone | HMDB | | 25-Methyldolichosterone | HMDB |
|
|---|
| Chemical Formula | C29H48O5 |
|---|
| Average Molecular Mass | 476.688 g/mol |
|---|
| Monoisotopic Mass | 476.350 g/mol |
|---|
| CAS Registry Number | 111618-87-2 |
|---|
| IUPAC Name | 14-(3,4-dihydroxy-6,6-dimethyl-5-methylideneheptan-2-yl)-4,5-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-8-one |
|---|
| Traditional Name | 14-(3,4-dihydroxy-6,6-dimethyl-5-methylideneheptan-2-yl)-4,5-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-8-one |
|---|
| SMILES | CC(C(O)C(O)C(=C)C(C)(C)C)C1CCC2C3CC(=O)C4CC(O)C(O)CC4(C)C3CCC12C |
|---|
| InChI Identifier | InChI=1S/C29H48O5/c1-15(25(33)26(34)16(2)27(3,4)5)18-8-9-19-17-12-22(30)21-13-23(31)24(32)14-29(21,7)20(17)10-11-28(18,19)6/h15,17-21,23-26,31-34H,2,8-14H2,1,3-7H3 |
|---|
| InChI Key | HAOQPQZVDMAOKT-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tetrahydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears four hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Tetrahydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Ergosterol-skeleton
- Ergostane-skeleton
- Tetrahydroxy bile acid, alcohol, or derivatives
- Ecdysteroid
- 23-hydroxysteroid
- 22-hydroxysteroid
- 3-hydroxysteroid
- 2-hydroxysteroid
- Hydroxysteroid
- Oxosteroid
- 6-oxosteroid
- Cyclic alcohol
- Ketone
- Secondary alcohol
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Organic oxygen compound
- Organooxygen compound
- Carbonyl group
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-1507900000-897e16ccbea7ecb07da8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-004i-2210029000-30690aab9742d3c68536 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06vi-0102900000-2fd81e8b2a091a103605 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4908600000-5884bd376a7478af5388 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-4903000000-6cf175c82182fbb9e7b8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0001900000-60025f5dbcc3fecbecba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9208700000-89a36f122832868194d9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9104000000-b96e458fdd73a3345812 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06ur-1019400000-6c5a3de93160cfe40425 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-07w9-1019100000-104c888a3346d4106002 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9501000000-950dac189be4f90068b8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0001900000-86a4f613459c2b1bffae | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-1105900000-992b4544e6eac49e2a84 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0009400000-ad283d56f2ffd21611a1 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039443 |
|---|
| FooDB ID | FDB019040 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000196 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 169366 |
|---|
| PubChem Compound ID | 13991214 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|