| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:41:20 UTC |
|---|
| Update Date | 2016-11-09 01:20:47 UTC |
|---|
| Accession Number | CHEM032771 |
|---|
| Identification |
|---|
| Common Name | ent-15,16-Epoxy-1(10),13(16),14-halimatrien-19-oic acid |
|---|
| Class | Small Molecule |
|---|
| Description | ent-15,16-Epoxy-1(10),13(16),14-halimatrien-19-oic acid is found in fruits. ent-15,16-Epoxy-1(10),13(16),14-halimatrien-19-oic acid is isolated (as Me ester) from seed-pod resin of Hymenaea courbaril (copinol). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
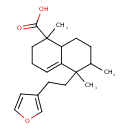
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| ent-15,16-Epoxy-1(10),13(16),14-halimatrien-19-Oate | Generator | | 5-[2-(Furan-3-yl)ethyl]-1,5,6-trimethyl-1,2,3,5,6,7,8,8a-octahydronaphthalene-1-carboxylate | HMDB |
|
|---|
| Chemical Formula | C20H28O3 |
|---|
| Average Molecular Mass | 316.435 g/mol |
|---|
| Monoisotopic Mass | 316.204 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 5-[2-(furan-3-yl)ethyl]-1,5,6-trimethyl-1,2,3,5,6,7,8,8a-octahydronaphthalene-1-carboxylic acid |
|---|
| Traditional Name | 5-[2-(furan-3-yl)ethyl]-1,5,6-trimethyl-2,3,6,7,8,8a-hexahydronaphthalene-1-carboxylic acid |
|---|
| SMILES | CC1CCC2C(=CCCC2(C)C(O)=O)C1(C)CCC1=COC=C1 |
|---|
| InChI Identifier | InChI=1S/C20H28O3/c1-14-6-7-17-16(5-4-10-20(17,3)18(21)22)19(14,2)11-8-15-9-12-23-13-15/h5,9,12-14,17H,4,6-8,10-11H2,1-3H3,(H,21,22) |
|---|
| InChI Key | ULDQVWIHFPQAFF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as colensane and clerodane diterpenoids. These are diterpenoids with a structure based on the clerodane or the colensane skeleton. Clerodanes arise from labdanes by two methyl migrations. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Colensane and clerodane diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Clerodane diterpenoid
- Heteroaromatic compound
- Furan
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fa9-2191000000-153b5b674960513c9046 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-7097000000-82a67af4238d10631f7e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0198000000-eb2ad7302d0128753366 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fya-1391000000-edc3247206489387a498 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kbr-2950000000-9b3bcddfc3e226d21e41 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0049000000-b0204cf858d612f2b453 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xr-0094000000-2cd0d99af9b72a39e5e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00ll-1090000000-d571cc306a9242559d01 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gi0-0491000000-d04d0d4bb7e59a66ed16 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fmr-2960000000-19cc3019dd6fb053496b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0lk9-8951000000-1dbf36eb55c9029f6704 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0009000000-d337ec43015bf5d00be6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0069000000-7f5eb8d0caaf1eeb7809 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-1192000000-62fbf132590ce74492ae | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039442 |
|---|
| FooDB ID | FDB019038 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014800 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752649 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|