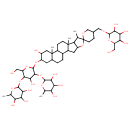| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:39:19 UTC |
|---|
| Update Date | 2016-11-09 01:20:46 UTC |
|---|
| Accession Number | CHEM032727 |
|---|
| Identification |
|---|
| Common Name | Tuberoside L |
|---|
| Class | Small Molecule |
|---|
| Description | Tuberoside L is found in onion-family vegetables. Tuberoside L is a constituent of Chinese chives seeds (Allium tuberosum). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C51H84O23 |
|---|
| Average Molecular Mass | 1065.199 g/mol |
|---|
| Monoisotopic Mass | 1064.540 g/mol |
|---|
| CAS Registry Number | 373647-11-1 |
|---|
| IUPAC Name | 2-{[4-hydroxy-2-(hydroxymethyl)-5-[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]-6-[7',9',13'-trimethyl-5-({[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}methyl)-5'-oxaspiro[oxane-2,6'-pentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icosane]-15'-oloxy]oxan-3-yl]oxy}-6-methyloxane-3,4,5-triol |
|---|
| Traditional Name | 2-{[4-hydroxy-2-(hydroxymethyl)-5-[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]-6-[7',9',13'-trimethyl-5-({[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}methyl)-5'-oxaspiro[oxane-2,6'-pentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icosane]-15'-oloxy]oxan-3-yl]oxy}-6-methyloxane-3,4,5-triol |
|---|
| SMILES | CC1C2C(CC3C4CCC5CC(OC6OC(CO)C(OC7OC(C)C(O)C(O)C7O)C(O)C6OC6OC(C)C(O)C(O)C6O)C(O)CC5(C)C4CCC23C)OC11CCC(COC2OC(CO)C(O)C(O)C2O)CO1 |
|---|
| InChI Identifier | InChI=1S/C51H84O23/c1-19-32-29(74-51(19)11-8-22(18-66-51)17-65-45-39(61)38(60)35(57)30(15-52)70-45)13-26-24-7-6-23-12-28(27(54)14-50(23,5)25(24)9-10-49(26,32)4)69-48-44(73-47-41(63)37(59)34(56)21(3)68-47)42(64)43(31(16-53)71-48)72-46-40(62)36(58)33(55)20(2)67-46/h19-48,52-64H,6-18H2,1-5H3 |
|---|
| InChI Key | JNVASJAUPSEUEW-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as steroidal saponins. These are saponins in which the aglycone moiety is a steroid. The steroidal aglycone is usually a spirostane, furostane, spirosolane, solanidane, or curcubitacin derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroidal glycosides |
|---|
| Direct Parent | Steroidal saponins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Steroidal saponin
- Triterpenoid
- Spirostane skeleton
- Oligosaccharide
- 2-hydroxysteroid
- Hydroxysteroid
- Glycosyl compound
- O-glycosyl compound
- Ketal
- Oxane
- Cyclic alcohol
- Tetrahydrofuran
- Secondary alcohol
- Organoheterocyclic compound
- Oxacycle
- Acetal
- Polyol
- Organooxygen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Primary alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uxs-4120123419-c20b72394a360441cddb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0w93-1211334906-b005719fe86164544b97 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0mb9-3211224903-1d4fd03e6b3bcba8f187 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9430012264-4052e4eba9a6340aba5d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fs-6900114233-2455577d2b8fdfccb926 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-08fr-4920014201-a6b4466da891ec7b6fbe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-016s-9600010004-52bcb6577b661359c64c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-8900040707-847b3ea170e83a39a363 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-057i-9410110014-fd95249eea1c2f4a70d8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-9100000011-cf7ca0269fcc4d2d42c9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fu-9300000002-60756694b273f2c53407 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05mo-9300000712-5cbadfcf4070a6fe2e48 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0039397 |
|---|
| FooDB ID | FDB018969 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752635 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|