| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:11:54 UTC |
|---|
| Update Date | 2016-11-09 01:19:25 UTC |
|---|
| Accession Number | CHEM032180 |
|---|
| Identification |
|---|
| Common Name | Solanocardinol |
|---|
| Class | Small Molecule |
|---|
| Description | Alkaloid from roots of Lycopersicon pimpinellifolium (currant tomato). 22-Isopimpifolidine is found in garden tomato. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
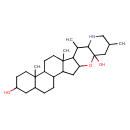
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Pimpifolidine | HMDB |
|
|---|
| Chemical Formula | C27H45NO3 |
|---|
| Average Molecular Mass | 431.651 g/mol |
|---|
| Monoisotopic Mass | 431.340 g/mol |
|---|
| CAS Registry Number | 138665-46-0 |
|---|
| IUPAC Name | 10,14,16,20-tetramethyl-23-oxa-18-azahexacyclo[12.11.0.0²,¹¹.0⁵,¹⁰.0¹⁵,²⁴.0¹⁷,²²]pentacosane-7,22-diol |
|---|
| Traditional Name | 10,14,16,20-tetramethyl-23-oxa-18-azahexacyclo[12.11.0.0²,¹¹.0⁵,¹⁰.0¹⁵,²⁴.0¹⁷,²²]pentacosane-7,22-diol |
|---|
| SMILES | CC1CNC2C(C)C3C(CC4C5CCC6CC(O)CCC6(C)C5CCC34C)OC2(O)C1 |
|---|
| InChI Identifier | InChI=1S/C27H45NO3/c1-15-13-27(30)24(28-14-15)16(2)23-22(31-27)12-21-19-6-5-17-11-18(29)7-9-25(17,3)20(19)8-10-26(21,23)4/h15-24,28-30H,5-14H2,1-4H3 |
|---|
| InChI Key | ZHFCFFSSVSIEIR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | This compound belongs to the class of organic compounds known as solanocapsine-type alkaloids. These are steroidal alkaloids with a structure that is characterized by a docosahydronaphth[2,1:4',5']indene ring system fused to a pyrano[3,2-b]pyridine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroidal alkaloids |
|---|
| Direct Parent | Solanocapsine-type alkaloids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Solanocapsine skeleton
- 23-hydroxysteroid
- 3-hydroxysteroid
- Hydroxysteroid
- Azasteroid
- Alkaloid or derivatives
- Oxane
- Piperidine
- Cyclic alcohol
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Secondary amine
- Azacycle
- Organoheterocyclic compound
- Secondary aliphatic amine
- Alcohol
- Organopnictogen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-11pi-1424900000-eb011db8396e14ea3c7c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-03di-5122290000-9506c1f6540e75dd506f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0011900000-36a85dd75a2ec3d6d5ff | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0230-6069600000-b1ba434fb5005af81a78 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9034000000-172eb6acd6f67d327fb6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0100900000-b949c409cb83fbdb31f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-1401900000-8d72889143988ad13c49 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-46142027663ee3676b4a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000900000-f1698e1bad247bcaa16d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0000900000-541706e9269bc6ebe443 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01si-0001900000-10ce5a6f3c1c22e03757 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0001900000-4cd9ae3fdf127416787b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-0223900000-171d8962e9bb5d019944 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0c00-3912000000-e177ac64c5ddfc367e15 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038836 |
|---|
| FooDB ID | FDB020574 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 73802037 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|