| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:09:36 UTC |
|---|
| Update Date | 2016-11-09 01:19:24 UTC |
|---|
| Accession Number | CHEM032127 |
|---|
| Identification |
|---|
| Common Name | 1,8-Heptadecadiene-4,6-diyne-3,10-diol |
|---|
| Class | Small Molecule |
|---|
| Description | 1,8-Heptadecadiene-4,6-diyne-3,10-diol is found in tea. 1,8-Heptadecadiene-4,6-diyne-3,10-diol is isolated from Panax quinquefolium (American ginseng). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
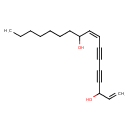
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,8-HDDDD | HMDB | | 8(e)-Heptadeca-1,8-dien-4,6-diyn-3,10-diol | HMDB | | Seselidiol | HMDB | | Heptadeca-1,8-diene-4,6-diyne-3,10-diol | MeSH |
|
|---|
| Chemical Formula | C17H24O2 |
|---|
| Average Molecular Mass | 260.371 g/mol |
|---|
| Monoisotopic Mass | 260.178 g/mol |
|---|
| CAS Registry Number | 63910-76-9 |
|---|
| IUPAC Name | (8Z)-heptadeca-1,8-dien-4,6-diyne-3,10-diol |
|---|
| Traditional Name | (8Z)-heptadeca-1,8-dien-4,6-diyne-3,10-diol |
|---|
| SMILES | CCCCCCCC(O)\C=C/C#CC#CC(O)C=C |
|---|
| InChI Identifier | InChI=1S/C17H24O2/c1-3-5-6-7-11-14-17(19)15-12-9-8-10-13-16(18)4-2/h4,12,15-19H,2-3,5-7,11,14H2,1H3/b15-12- |
|---|
| InChI Key | DSVMWGREWREVQQ-QINSGFPZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain fatty alcohols. These are fatty alcohols that have an aliphatic tail of 13 to 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohols |
|---|
| Direct Parent | Long-chain fatty alcohols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Long chain fatty alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03ec-5920000000-eb7064c25041144fb90d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00tr-9017000000-dd22962ee0fe08ec5b38 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-0190000000-534bf2aa550119896d12 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p6-9860000000-d6396f7a97b5a26363a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9300000000-8678a2103c3790851e2c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-2007e2dced24c5607ca6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-2290000000-a11d89d802ca7205104d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-7920000000-00a84a074c7579033e13 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-052f-0090000000-3b1cbfe05ce6fa050cc8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4l-1590000000-60b698263ad9e409547a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0odi-4910000000-a3454d3965dac4e9cd8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0690000000-37c5569af5cf5c3ffcdd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-5890000000-6e3aa4a8b949b55320ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01di-9700000000-a9ae8163065385cd6f4c | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038781 |
|---|
| FooDB ID | FDB018200 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00031489 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4943085 |
|---|
| ChEBI ID | 66471 |
|---|
| PubChem Compound ID | 6438621 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|