| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:08:07 UTC |
|---|
| Update Date | 2016-11-09 01:19:24 UTC |
|---|
| Accession Number | CHEM032092 |
|---|
| Identification |
|---|
| Common Name | Glyuranolide |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Glycyrrhiza uralensis (Chinese licorice). Glyuranolide is found in herbs and spices. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
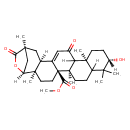
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Methyl 3-hydroxy-11-oxo-12-oleanen-29,22a-olid-27-Oate | HMDB | | Methyl (1S,2R,5R,6R,11S,14S,19S,21S)-11-hydroxy-2,6,10,10,14,21-hexamethyl-16,22-dioxo-23-oxahexacyclo[19.2.1.0²,¹⁹.0⁵,¹⁸.0⁶,¹⁵.0⁹,¹⁴]tetracos-17-ene-5-carboxylic acid | Generator | | 3,22-Dihydroxy-11-oxo-delta(12)-oleanene-27-alpha-methoxycarbonyl-29-Oic acid | MeSH | | Glyuranolide | MeSH |
|
|---|
| Chemical Formula | C31H44O6 |
|---|
| Average Molecular Mass | 512.678 g/mol |
|---|
| Monoisotopic Mass | 512.314 g/mol |
|---|
| CAS Registry Number | 123914-44-3 |
|---|
| IUPAC Name | methyl (1S,2R,5R,6R,11S,14S,19S,21S)-11-hydroxy-2,6,10,10,14,21-hexamethyl-16,22-dioxo-23-oxahexacyclo[19.2.1.0²,¹⁹.0⁵,¹⁸.0⁶,¹⁵.0⁹,¹⁴]tetracos-17-ene-5-carboxylate |
|---|
| Traditional Name | methyl (1S,2R,5R,6R,11S,14S,19S,21S)-11-hydroxy-2,6,10,10,14,21-hexamethyl-16,22-dioxo-23-oxahexacyclo[19.2.1.0²,¹⁹.0⁵,¹⁸.0⁶,¹⁵.0⁹,¹⁴]tetracos-17-ene-5-carboxylate |
|---|
| SMILES | [H][C@@]12C[C@@]3(C)C[C@H](OC3=O)[C@]1(C)CC[C@]1(C(=O)OC)C2=CC(=O)C2[C@@]3(C)CC[C@H](O)C(C)(C)C3CC[C@@]12C |
|---|
| InChI Identifier | InChI=1S/C31H44O6/c1-26(2)20-8-11-30(6)23(29(20,5)10-9-21(26)33)19(32)14-17-18-15-27(3)16-22(37-24(27)34)28(18,4)12-13-31(17,30)25(35)36-7/h14,18,20-23,33H,8-13,15-16H2,1-7H3/t18-,20?,21-,22-,23?,27-,28+,29-,30+,31+/m0/s1 |
|---|
| InChI Key | MKDSBDQLSLPNOQ-OZFAJWBBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- 19-oxosteroid
- Oxosteroid
- Steroid
- Caprolactone
- Cyclohexenone
- Oxepane
- Dicarboxylic acid or derivatives
- Gamma butyrolactone
- Cyclic alcohol
- Methyl ester
- Tetrahydrofuran
- Carboxylic acid ester
- Ketone
- Lactone
- Secondary alcohol
- Organoheterocyclic compound
- Carboxylic acid derivative
- Oxacycle
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uyj-0201900000-2af370945107e9ee6142 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00xu-1131290000-e9cb9681d63512bd7421 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ot-0000930000-bbcce3f44f49a5edeba0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ot-0100900000-3ee1634c5674f58fab8d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00m0-2324900000-21d102874cad1084a828 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000690000-3a72b54f4c0de145b299 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xr-0000930000-d3ad3beedea0497db536 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ktr-2000900000-f087ac1072654348d3b0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-0b08871528dc53be3c09 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0000290000-0eaf6159be15b68de7cb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ikj-2000910000-7f74b0e46504c60a2343 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000590000-9acf1efeaeef01fe7d34 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000930000-6732dc10adddc7704308 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udj-5836900000-81b6327e8ed9fe98e08c | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038745 |
|---|
| FooDB ID | FDB018156 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4476521 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752447 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|