| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:07:46 UTC |
|---|
| Update Date | 2016-11-09 01:19:24 UTC |
|---|
| Accession Number | CHEM032083 |
|---|
| Identification |
|---|
| Common Name | (3S,5R,6R,7E)-3,5,6-Trihydroxy-7-megastigmen-9-one |
|---|
| Class | Small Molecule |
|---|
| Description | (3S,5R,6R,7E)-3,5,6-Trihydroxy-7-megastigmen-9-one is found in herbs and spices. (3S,5R,6R,7E)-3,5,6-Trihydroxy-7-megastigmen-9-one is a constituent of New Zealand thyme honey (Thymus vulgaris). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
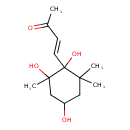
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C13H22O4 |
|---|
| Average Molecular Mass | 242.311 g/mol |
|---|
| Monoisotopic Mass | 242.152 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (3E)-4-(1,2,4-trihydroxy-2,6,6-trimethylcyclohexyl)but-3-en-2-one |
|---|
| Traditional Name | (3E)-4-(1,2,4-trihydroxy-2,6,6-trimethylcyclohexyl)but-3-en-2-one |
|---|
| SMILES | CC(=O)\C=C\C1(O)C(C)(C)CC(O)CC1(C)O |
|---|
| InChI Identifier | InChI=1S/C13H22O4/c1-9(14)5-6-13(17)11(2,3)7-10(15)8-12(13,4)16/h5-6,10,15-17H,7-8H2,1-4H3/b6-5+ |
|---|
| InChI Key | QASOACWKTAXFSE-AATRIKPKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sesquiterpenoids. These are terpenes with three consecutive isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sesquiterpenoid
- Megastigmane sesquiterpenoid
- Cyclofarsesane sesquiterpenoid
- Ionone derivative
- Cyclohexanol
- Cyclitol or derivatives
- Acryloyl-group
- Cyclic alcohol
- Enone
- Alpha,beta-unsaturated ketone
- Tertiary alcohol
- Ketone
- Secondary alcohol
- Polyol
- Organooxygen compound
- Organic oxide
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002e-5930000000-cecf9f3b0925fdb8d534 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-002f-5115900000-dfb719fbe5ba5faf547a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004l-0190000000-3c1f704e0fa28ef04ac4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-2390000000-d89370be9a22de9048fd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9410000000-eae65e535911e5cd63b6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0290000000-50f3b4c1aa659fb0dfd9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-4690000000-035377d8ce4535c9d2ee | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0592-8910000000-142c54cd7d405f1eecaf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0490000000-fc77103efd0e16d722c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-0900000000-2e20bede7b265f622da8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0900000000-954704a2782d55250133 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05ru-0960000000-14d1dd0add97b1c13dec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-7930000000-a3755251020ef166a53c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6x-9300000000-7188e1c0ae1a49054a33 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038736 |
|---|
| FooDB ID | FDB018147 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 31014626 |
|---|
| ChEBI ID | 168487 |
|---|
| PubChem Compound ID | 51136538 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|