| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:07:43 UTC |
|---|
| Update Date | 2016-11-09 01:19:24 UTC |
|---|
| Accession Number | CHEM032081 |
|---|
| Identification |
|---|
| Common Name | Macrocarpal E |
|---|
| Class | Small Molecule |
|---|
| Description | Macrocarpal E is a constituent of Eucalyptus globulus (Tasmanian blue gum). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
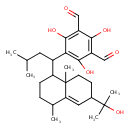
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C28H40O6 |
|---|
| Average Molecular Mass | 472.614 g/mol |
|---|
| Monoisotopic Mass | 472.282 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 2,4,6-trihydroxy-5-{1-[6-(2-hydroxypropan-2-yl)-4,8a-dimethyl-1,2,3,4,6,7,8,8a-octahydronaphthalen-1-yl]-3-methylbutyl}benzene-1,3-dicarbaldehyde |
|---|
| Traditional Name | 2,4,6-trihydroxy-5-{1-[6-(2-hydroxypropan-2-yl)-4,8a-dimethyl-2,3,4,6,7,8-hexahydro-1H-naphthalen-1-yl]-3-methylbutyl}benzene-1,3-dicarbaldehyde |
|---|
| SMILES | CC(C)CC(C1CCC(C)C2=CC(CCC12C)C(C)(C)O)C1=C(O)C(C=O)=C(O)C(C=O)=C1O |
|---|
| InChI Identifier | InChI=1S/C28H40O6/c1-15(2)11-18(23-25(32)19(13-29)24(31)20(14-30)26(23)33)21-8-7-16(3)22-12-17(27(4,5)34)9-10-28(21,22)6/h12-18,21,31-34H,7-11H2,1-6H3 |
|---|
| InChI Key | XJNGQIYBMXBCRU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as eudesmane, isoeudesmane or cycloeudesmane sesquiterpenoids. These are sesquiterpenoids with a structure based on the eudesmane skeleton. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Eudesmane, isoeudesmane or cycloeudesmane sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Eudesmane, isoeudesmane or cycloeudesmane sesquiterpenoid
- Acylphloroglucinol derivative
- Benzenetriol
- Hydroxybenzaldehyde
- Phloroglucinol derivative
- Benzaldehyde
- Benzoyl
- Aryl-aldehyde
- Phenol
- Monocyclic benzene moiety
- Benzenoid
- Vinylogous acid
- Tertiary alcohol
- Polyol
- Organooxygen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Organic oxide
- Aldehyde
- Aromatic homopolycyclic compound
|
|---|
| Molecular Framework | Aromatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-7043900000-22e3e56bcf2b5e0a0d7a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00e9-3112019000-6365d73b9406f855e6d7 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-0010900000-c6ba33365147f4db7509 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-2043900000-d06f43b9833fdfe2263d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pi9-4069300000-ab132233e0d75c630063 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-18b45f07fb54edca02c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0h9r-0210900000-5a56917b2782d8fa7c23 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01rx-5976600000-a80618b6bb3702ae2a39 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fs-1043900000-104f6ee8bfc3f70123f7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fmi-0393300000-a7d554e51f16f15bc42a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-2900000000-2942b08a1653070f64bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-025afde3cd0f17166439 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0kmi-3610900000-8898b0edb926ff81c21b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fef-6936300000-a84d99c7a61ede554ad3 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038734 |
|---|
| FooDB ID | FDB018145 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 400196 |
|---|
| ChEBI ID | 175708 |
|---|
| PubChem Compound ID | 454461 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|